Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 59(4); 2025 > Article
-
Original Article
AMACR is a highly sensitive and specific immunohistochemical marker for diagnosing prostate cancer on biopsy: a systematic review and meta-analysis -
Johannes Cansius Prihadi1,2,*, Stevan Kristian Lionardi2,3,*
 , Nicolas Daniel Widjanarko3
, Nicolas Daniel Widjanarko3 , Steven Alvianto4
, Steven Alvianto4 , Fransiskus Xaverius Rinaldi5
, Fransiskus Xaverius Rinaldi5 , Archie Fontana Iskandar6
, Archie Fontana Iskandar6
-
Journal of Pathology and Translational Medicine 2025;59(4):235-248.
DOI: https://doi.org/10.4132/jptm.2025.04.16
Published online: July 3, 2025
1Department of Urology, Atma Jaya Hospital, Jakarta, Indonesia
2Department of Surgery, Division of Urology, School of Medicine and Health Science, Atma Jaya Catholic University of Indonesia, Jakarta, Indonesia
3Fatima Hospital, Ketapang, West Kalimantan, Indonesia
4School of Medicine and Health Science, Atma Jaya Catholic University of Indonesia, Jakarta, Indonesia
5Santo Antonius Jopu Hospital, Ende, East Nusa Tenggara, Indonesia
6Cimacan General Regional Hospital, Cianjur, West Java, Indonesia
- Corresponding Author: Stevan Kristian Lionardi Department of Surgery, Division of Urology, School of Medicine and Health Science, Atma Jaya Catholic University of Indonesia, Jl. Pluit Raya No. 2, Penjaringan, North Jakarta, DKI Jakarta, 14440, Indonesia Tel: +62-81289374200, Fax: +62-81289374200, E-mail: stevanlionardi15@gmail.com
- *Johannes Cansius Prihadi and Stevan Kristian Lionardi contributed equally to this work.
© The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Alpha-methylacyl-CoA racemase (AMACR) is the preferred biomarker for distinguishing malignant from benign glands in prostate biopsies, showing high sensitivity and specificity for prostate cancer. A meta-analysis of immunohistochemistry (IHC) for AMACR is essential to further assess its diagnostic accuracy across diverse sample sources.
-
Methods
- A systematic search of databases including MEDLINE, ScienceDirect, ProQuest, Google Scholar, and the Cochrane Library was performed, focusing on studies of AMACR to diagnose prostate cancer, particularly in biopsy samples analyzed through IHC over the last 20 years. Quality of studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 tool, followed by a meta-analysis of regions and subgroups to calculate summary estimates of diagnostic test accuracy.
-
Results
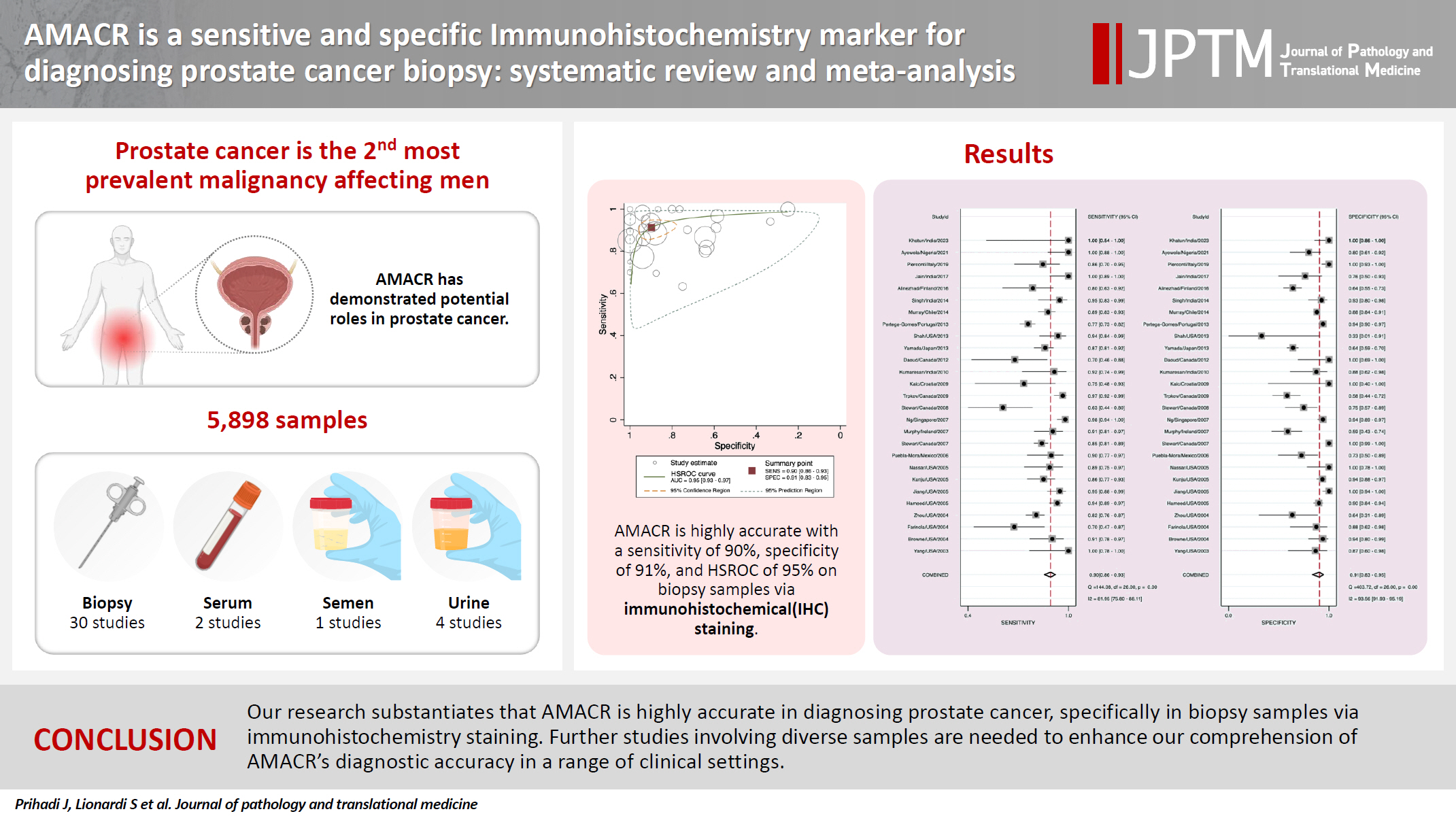
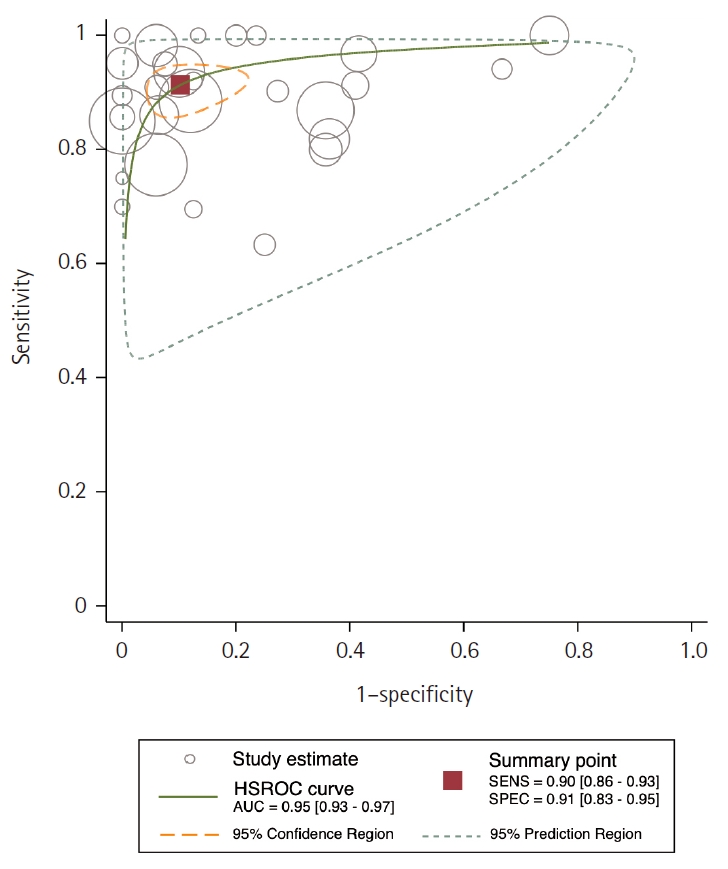
- In the final analysis, 37 studies, with a pooled size of 5,898 samples, were included from the examination of 94 full-text papers. Among them, 27 studies with similar sample sources and testing methodologies underwent meta-analysis, yielding a combined sensitivity estimate of 0.90 (95% confidence interval [CI], 0.86 to 0.93) and specificity of 0.91 (95% CI, 0.83 to 0.95), both with significant heterogeneity (p < .01). The region beneath the hierarchical summary receiver operating characteristic curve was 0.95 (95% CI, 0.93 to 0.97), positive likelihood ratio was 9.6 (95% CI, 5.3 to 17.4), negative likelihood ratio was 0.11 (95% CI, 0.08 to 0.15), and diagnostic odds ratio was 88 (95% CI, 42 to 181).
-
Conclusions
- Our meta-analysis findings substantiate AMACR as a highly accurate tool for diagnosing prostate cancer, specifically in biopsy samples, via immunohistochemical staining. Further studies involving diverse samples are needed to enhance our understanding of the AMACR diagnostic accuracy in a range of clinical settings.
- Prostate cancer (PCa) ranks as the second most prevalent malignancy affecting men, with an estimated 1.4 million diagnoses in 2020 [1]. Digital rectal examinations and prostate-specific antigen (PSA) tests have become standard practices for early identification of PCa. If an individual has elevated PSA values, additional imaging and biopsy procedures are recommended [2]. Among individuals categorized as low risk on magnetic resonance imaging results, 11 to 28 of every 100 have clinically significant cancer.
- Underdiagnosed PCa may result in delayed initiation of early treatment, potentially leading to significant adverse events for patients [3]. As PCa may not be distinguishable from benign prostate glands, its identification can occasionally present a diagnostic challenge [4]. Therefore, to augment the discriminatory capability of hematoxylin and eosin (H&E) staining in distinguishing between small foci of benign and malignant glands and numerous complex cases in which the pathologist exhibits no confidence in diagnosing solely based on H&E staining [5], immunohistochemical (IHC) markers of PCa are used as supplementary tools to reinforce critical diagnostic decisions, offering heightened diagnostic accuracy [3].
- Alpha-methylacyl-CoA racemase (AMACR) or P504S is an enzyme critical for stereochemical inversion in the metabolic pathways of several proteins and pharmaceutical agents. Located in both peroxisomes and mitochondria, AMACR has been shown to include splice variants with potential roles in cancer development, particularly PCa [6].
- While PSA serves as an important initial clinical biomarker, elevated levels still necessitate a pathological biopsy to confirm PCa and allow further classification through Gleason scoring. Differentiating glands with atypical, bland-appearing features from those with PCa is very difficult, particularly for cases with low Gleason scores (e.g., 3). Presently, prostate needle biopsy remains the gold standard for diagnosis of PCa; however, these are not entirely free of diagnostic inaccuracies. The current challenge faced by pathologists in the interpretation of prostate needle biopsy specimens is the increasing prevalence of samples containing a limited number of suspicious glands and minimal atypia [7]. The increasing prevalence of samples containing a limited number of suspicious glands and minimal atypia may be attributed to improved biopsy techniques, more widespread PSA screening, and enhanced pathological scrutiny. In this context, AMACR has diagnostic utility in diagnosis of PCa. These differences may be due to variations in diagnostic methods and potential differences among ethnic and racial groups [8].
- Recent research has shown that AMACR has a high sensitivity and specificity for PCa compared to routine H&E staining in resolving suspicious lesions and excluding benign mimickers. Therefore, AMACR in PCa is a promising candidate as a diagnostic marker [9]. A previous meta-analysis conducted by Jiang et al. (2013) [3] observed that the expression of AMACR was associated with a significantly increased risk of PCa. However, its sensitivity and specificity in terms of diagnostic accuracy were not evaluated. Therefore, given the lack of a comprehensive systematic review and meta-analysis, we performed the first meta-analysis to determine the diagnostic test accuracy (DTA) of AMACR as an essential marker in PCa diagnosis.
INTRODUCTION
- Source and search strategy
- The current investigation was registered in PROSPERO under registration number CRD42024495450. Systematic exploration published from January 2003 until December 2023 was performed across five databases: MEDLINE, ScienceDirect, ProQuest, Google Scholar, and the Cochrane Library. Key search terms used were ‘PCa,’ ‘AMACR,’ and ‘biomarkers.’ Additionally, manual screening of reference lists from included studies and a snowballing method were implemented to identify useful studies (Supplementary Table S1).
- Selection criteria
- Studies were deemed eligible when they fulfilled the following criteria: (1) examination of AMACR and its association with PCa, (2) inclusion of patients without other types of cancer, (3) performance of testing among benign or biopsy-negative patients, (4) utilization of pathology as the reference standard, (5) provision of relevant data amenable to accurate extraction, and (5) non-randomized studies of interventions.
- Studies were considered ineligible if they were reviews, conference proceedings, book sections, commentaries, editorials, letters, abstract-only publications, or in vitro or in silico investigations or if they presented inseparable independent AMACR data using mixed or cocktail diagnostic modalities. We used a population, intervention, comparison, outcome, timing, setting, and study design (PICOTS-SD) framework for the systematic review (Supplementary Table S2). The excluded potentially relevant studies are documented in the supplementary material (Supplementary Table S3).
- Data extraction and quality assessment
- Search results were downloaded and compiled into Zotero, a review database. Subsequently, each study was screened by all authors. NDW, SA, FXR, and AFI independently conducted data extraction from all included studies. For validation purposes, JCP and SKL performed another round of data extraction independently on respective halves of the included studies. Any discrepancies regarding study inclusion were resolved through adjudication by JCP. The following data metrics were extracted into a spreadsheet program: first author; country; year; study title; source of sample; control source; and percentages of true-positive, false-positive, false-negative, and true-negative tests. Documented cases of high-grade prostatic intraepithelial neoplasia (HGPIN) or atypical lesions progressing to PCa were also extracted.
- Transurethral resection of the prostate, radical prostatectomy, or needle biopsies were used to retrieve biopsy samples from patients diagnosed with prostate cancer. IHC or real-time polymerase chain reaction (RT-PCR) was then used to evaluate the biopsy samples. Patient blood samples and urinary sediments following digital rectal examination were analyzed using RT-PCR. Additionally, some studies reported analysis of urine samples using a chemiluminescence detection system or enzyme-linked immunosorbent assay. Semen ejaculates from biopsy-proven PCa patients before radical prostatectomy were also occasionally used as specimens.
- We referenced guidelines outlined in the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) 2020 statement [10] for the literature selection process and used the comprehensive PRISMA 2020 checklist (Supplementary Table S4). Included studies were assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool [11]. Each author independently evaluated the studies, with a balanced workload. Any discrepancies were addressed through deliberation and consultation with JCP.
- Outcome measures
- Diagnostic accuracy was assessed based on sensitivity and specificity, positive and negative predictive values (PPV and NPV), positive and negative likelihood ratios (PLR and NLR), diagnostic odds ratio (DOR), and area under the curve (AUC) of AMACR. Specifically, outcomes related to AMACR were evaluated based on evaluation method, i.e., IHC staining, RT-PCR, echo chemiluminescence detection system (ECL-DS), or chemiluminescence immunoassay (CLIA) of samples obtained from biopsy, urine, serum, and/or semen samples.
- Statistical analysis
- Data were analyzed to calculate true-positive, false-negative, false-positive, and true-negative values. We generated a hierarchical summary receiver operating characteristic (HSROC) curve to evaluate diagnostic performance across studies because it accounts for variability in meta-analyses of DTA. In more detail, the HSROC model, a random-effects model, incorporates both within- and between-study variability to provide a more reliable summary of test accuracy, particularly in the presence of threshold effects [12].
- A bivariate mixed-effects regression was used to calculate combined PPV and NPV, along with their corresponding 95% confidence intervals (CIs). Region and subgroup analyses were also performed to explore variations in DTA. Heterogeneity was evaluated using the I2 statistical method, where I2 > 50% or a p-value <.05 was considered to signify significant heterogeneity [13]. Publication bias was assessed through Deek’s funnel plot, with p < .05 considered statistically significant.
- All statistical analyses were conducted using Cochrane Review Manager software ver. 5.4 and the Metandi [14] and MIDAS [15] modules of Stata software ver. 17.0 (Stata Corp., College Station, TX, USA) and Meta-disc ver. 2.0 [16]. Measurements with a p-value less than .05 were regarded as statistically significant.
MATERIALS AND METHODS
- Study selection and characteristics
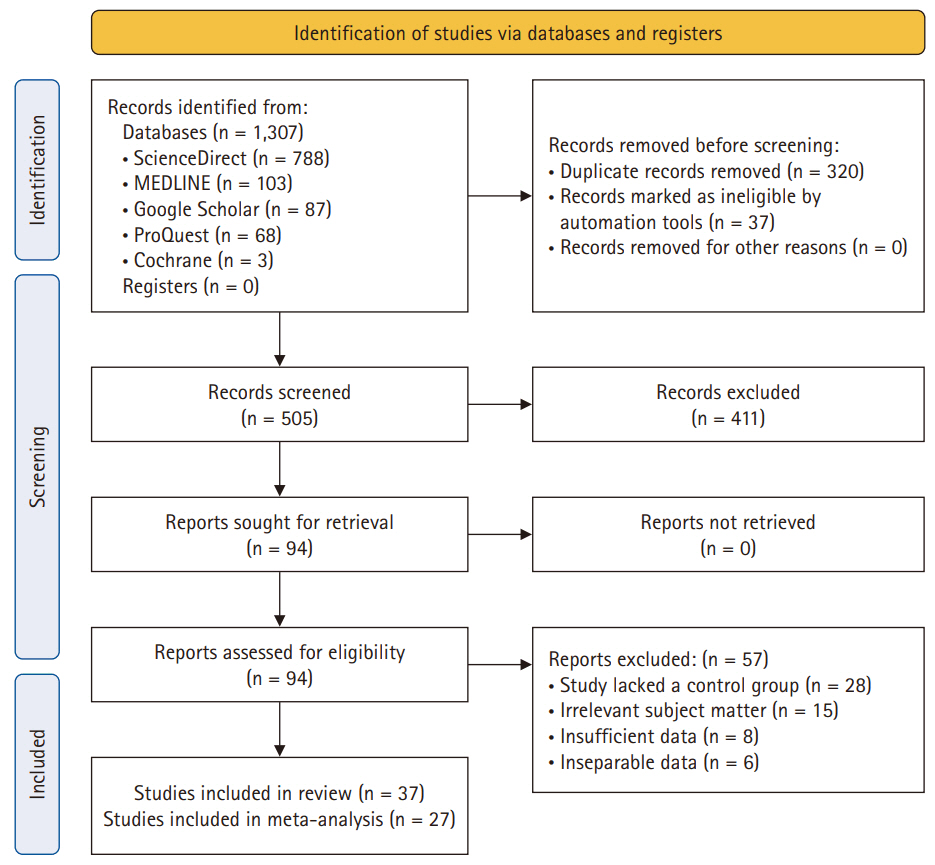
- After deduplication, a comprehensive screening process of 505 studies was performed. A subsequent assessment of titles and abstracts led to the exclusion of 411 studies, resulting in a thorough examination of the full text of 94 studies. Eventually, 37 studies were deemed suitable for inclusion in the analysis. Comprehensive details regarding the exclusion of full texts are provided in Fig. 1.
- Among the 31 investigations using biopsy samples, 27 [5,17-41] were analyzed with IHC and three using RT-PCR. Furthermore, urine, serum, and semen samples were used in four, two, and one study, respectively. There were 5,898 samples in total, ranging from 19 to 731 per study [5,17-52]. The majority of specimens (2,311) was collected from patients enrolled in 16 studies in the United States [5,17-23,34,42,43,45,46,48,49,51]. Four studies each were conducted in Canada [25,28,29,32] and India [31,38,41,52], while one to two studies each were conducted in other countries.
- Pathological testing served as the reference standard, with 32 studies using benign controls as the reference and five studies using biopsy-negative results. Thirteen studies had no conflicts of interest, while seven studies acknowledged academic conflicts of interest for grants or patents. Seventeen studies did not disclose conflicts of interest. Baseline characteristics of the included studies are outlined in Table 1.
- Progression of positive AMACR cases to PCa
- Across the included studies, seven [5,19,22,28,35,40,49] evaluated the progression of HGPIN or atypical cases with positive AMACR into PCa (Table 2). Stewart et al. [28] reported that HGPIN cases were five times more likely to progress to PCa, while Ayowole et al. [40] reported a 75% conversion rate of HGPIN to PCa in AMACR-positive cases. Additionally, nearly half of atypical cases also progressed to PCa [19,22,49]. Overall, the findings indicated that more than half of the cases with positive AMACR progressed to PCa.
- Research suggests that, when AMACR is absent, HGPIN or atypical cases are unlikely to develop into PCa. For example, Zhou et al. (2004) [19] followed 81 atypical cases and found all 39 AMACR-negative cases to be benign. Similarly, Ayowole et al. (2021) [40] explored the link between AMACR staining and histological diagnosis, showing that every AMACR-negative case was either normal prostate tissue (100%) or benign prostatic hyperplasia (14%), with none progressing to prostate cancer. These findings highlight the potential of AMACR as a reliable marker for distinguishing between benign and malignant prostate conditions.
- Quality assessment of studies
- Application of the QUADAS-2 tool for bias assessment revealed several potential areas of bias in the studies included. Thirty studies had a low risk of bias (ROB) in the patient selection domain, while the remaining seven studies were classified as high and unclear risk due to insufficient details regarding participant recruitment and exclusion criteria. ROB was rated high in 16 studies and unclear in 11 studies in terms of the index test domain, where most of the index test outcomes were obtained with knowledge of the reference standard test. No studies had a high ROB in the reference standard domain. Regarding the flow and time domains, only one study received a high ROB rating. All studies had low concern for applicability in all domains, as shown in Supplementary Fig. S1.
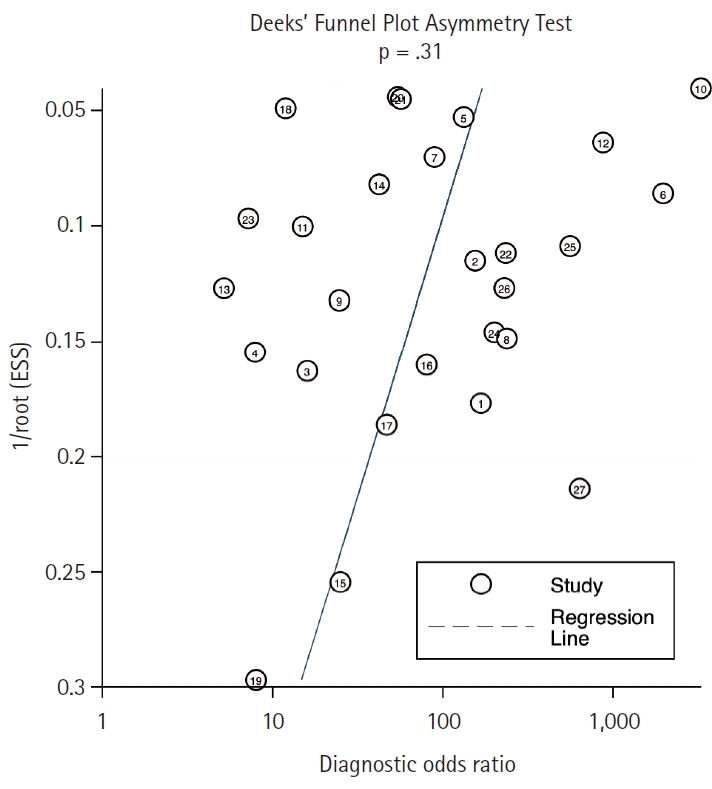
- Deek’s funnel-plot asymmetry test was employed to assess the presence of publication bias, yielding a non-significant result (p = .315), indicating the absence of such bias (Fig. 2).
- Diagnostic test accuracy
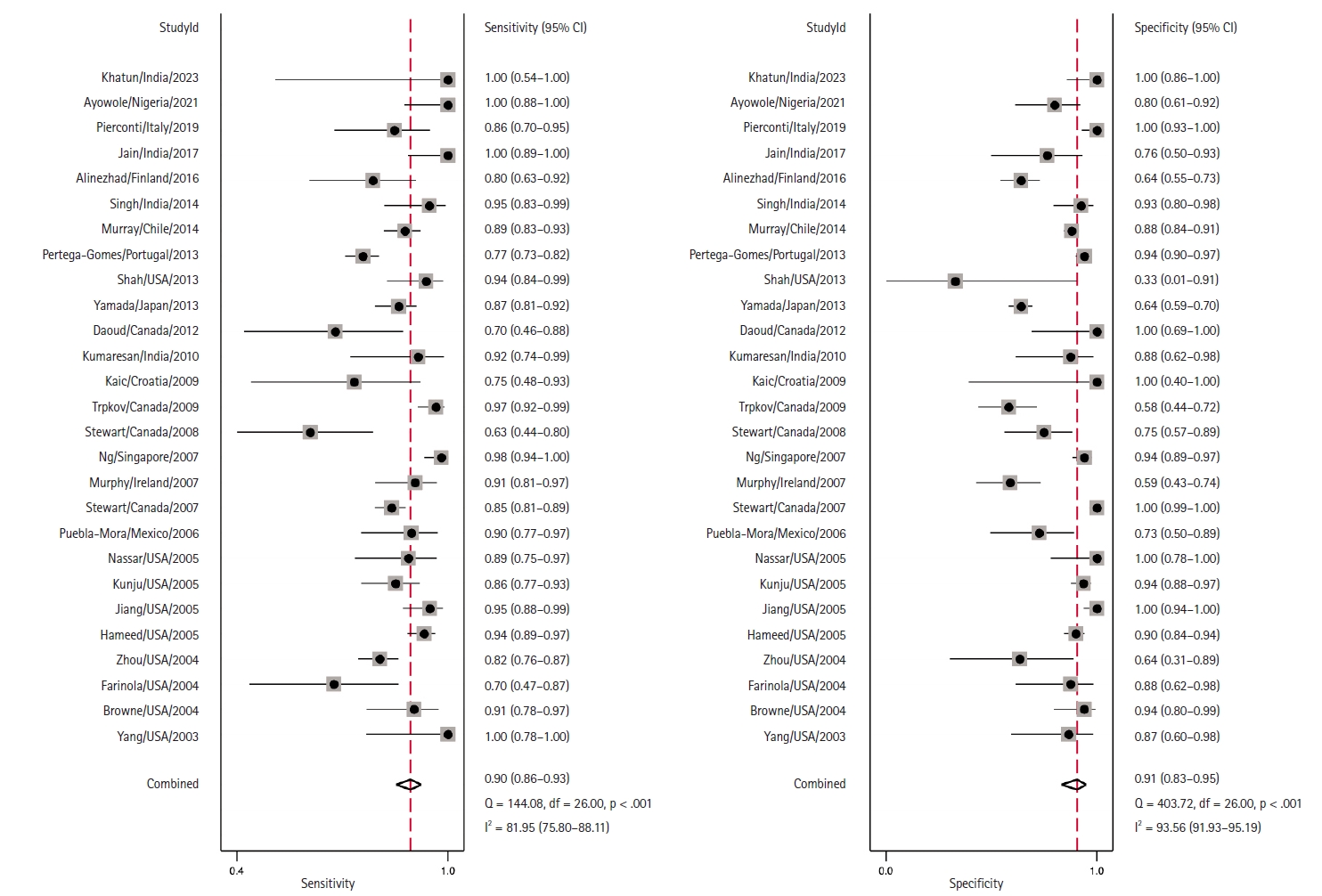
- Coupled Forest plots illustrating the sensitivity and specificity of each study are depicted in Fig. 3. The estimated combined sensitivity was 0.90 (95% CI, 0.86 to 0.93) with significant heterogeneity (I2 = 81.95, p < .01). Correspondingly, the combined specificity was 0.91 (95% CI, 0.83 to 0.95), also with significant heterogeneity (I2 = 93.56, p < .01).
- Area under the HSROC curve was determined to be 0.95 (95% CI, 0.93 to 0.97). Notably, the close alignment of the main solid curve to the top left indicates a high overall accuracy (Fig. 4). Moreover, the PLR was 9.6 (95% CI, 5.3 to 17.4), the NLR was 0.11 (95% CI, 0.08 to 0.15), and the DOR was 88 (95% CI, 42 to 181). A Fagan plot is presented in Supplementary Fig. S2.
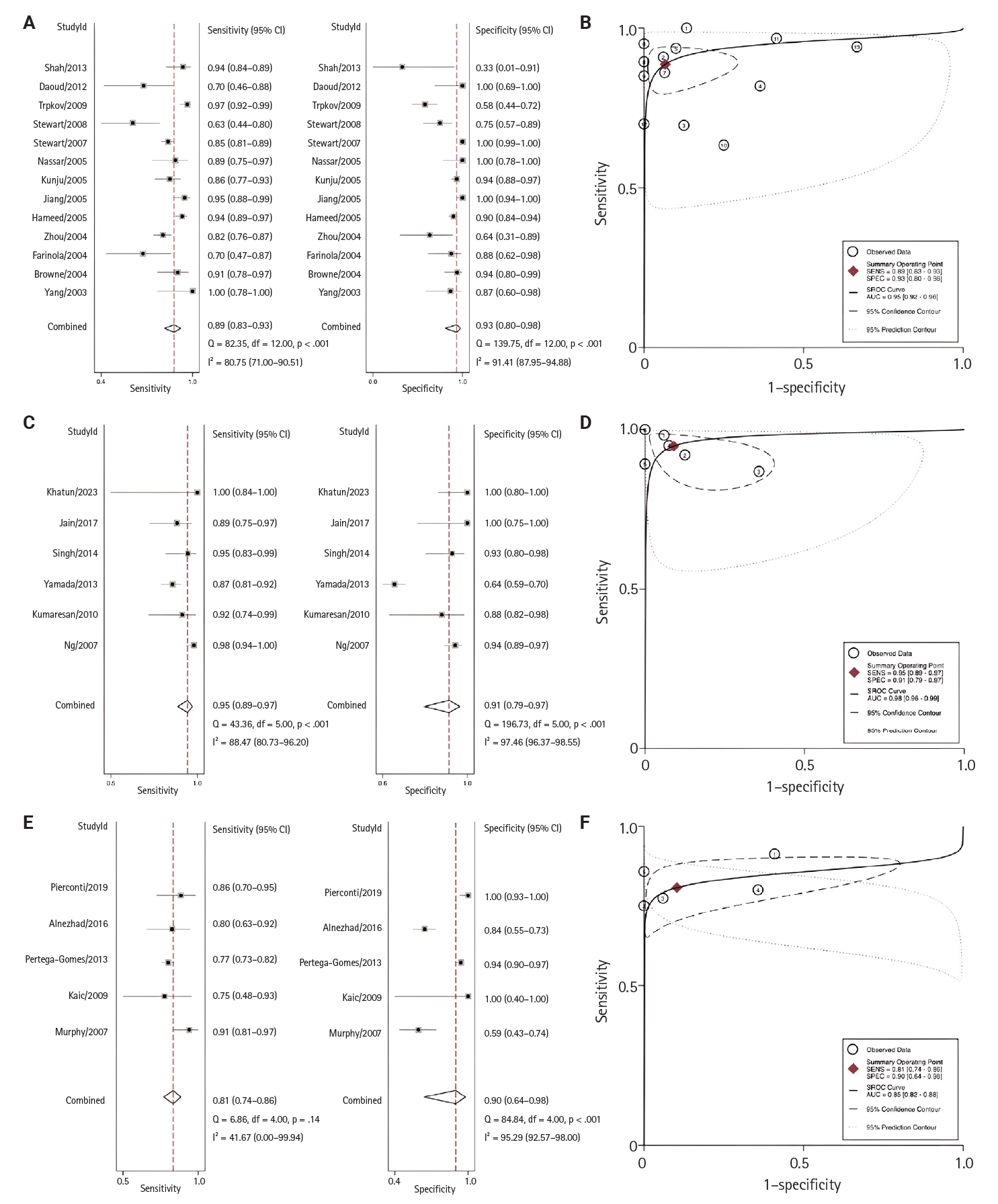
- Regional analyses were conducted according to the research country of origin (Fig. 5). From highest to lowest, the AUC for each region was 0.98 (95% CI, 0.96 to 0.98) for Asia, 0.95 (95% CI, 0.92 to 0.96) for North America, and 0.85 (95% CI, 0.82 to 0.88) for Europe.
- Results of subgroup analysis according to sample type are presented in Table 1. Rogers et al. [49], who used an echo CLIA of urine samples; Sroka et al. [50], who used CLIA of urine samples; and Etheridge et al. [51], who used CLIA analysis of semen samples, were grouped under ‘others’ due to the limited number of studies using these sample types.
RESULTS
- Summary of findings
- In this analysis, we evaluated the prospective diagnostic accuracy of AMACR for PCa diagnosis among participants attending healthcare facilities. We systematically reviewed and analyzed 37 observational studies regardless of methodology and found that AMACR had a sensitivity and specificity of at least 88% for diagnosis of PCa (Supplementary Table S4). In 27 studies with uniform sample sources (biopsy) and testing methodologies (IHC), a minimum sensitivity and specificity of 90% were attained. RT-PCR methods demonstrated a sensitivity of 83% and a specificity of 91%; however, in urine and serum samples, both sensitivity and specificity were significantly lower. Notably, these findings are based on limited data, as only 2–3 studies have evaluated RT-PCR performance for these sample types, limiting the generalizability of these results.
- Initially documented in 2000 in two adults exhibiting neurological disorders linked to its deficiency [53], AMACR has since been identified in numerous other neurological cases [54-56]. Elevated expression levels of AMACR have been observed in 32 types of human cancers, as identified by The Cancer Genome Atlas Program (TCGA) [57,58], including gastrointestinal [59,60], ovarian [61], renal [62], and liver [63] cancers. Notably, AMACR level was most highly elevated in PCa, with a 682-fold increase in mRNA levels compared to control samples [37]. AMACR also exhibits preferential overexpression in approximately 80%–100% of PCa identified through biopsy [64].
- In our meta-analysis, AMACR exhibited exceptional sensitivity and specificity as a diagnostic biomarker of PCa, with values of 90% and 91%, respectively, as shown in the Forest plot in Fig. 3. These findings suggest that AMACR is appropriate to use for risk stratification of PCa. Furthermore, the AUC of 0.95 underscores the assay's remarkable discriminatory power (Fig. 4). The Fagan nomogram (Supplementary Fig. S2) corroborated this exceptional accuracy by demonstrating a high PLR of 9.6 and a low NLR of 0.11. Collectively, these results indicate that AMACR is a highly accurate diagnostic tool for differentiating between patients with PCa and those with benign prostatic conditions.
- IHC of biopsy samples was the most widely used method to assess AMACR expression in the studies we reviewed, with higher diagnostic accuracy than other sample types. RT-PCR analysis, however, has its own advantages; for example, detection of AMACR expression in various prostatic secretions (urine, serum, semen). Automated RT-PCR analysis also helps reduce error rates in pathological diagnosis, particularly in centers without pathologists specialized in urological pathology [42].
- Although diagnostic accuracy was lower for urinary (sensitivity, 72%; specificity, 69%) and serum (sensitivity, 70%; specificity, 39%) samples, these types remain appealing as they can be collected using minimally invasive methods, especially from patients unable to undergo biopsy due to medical or other conditions. Further studies exploring prostatic secretions and serum in combination with PSA or other examinations could enhance the diagnostic accuracy of AMCAR in these sample types and aid in the development of non-invasive diagnostic protocols [65].
- This study evaluated the probability of progression from HGPIN and atypical lesions to PCa in cases positive for AMACR expression and found that this was high. AMACR is used to diagnose several tumor markers and their precursor lesions, including PCa [66]. Our findings indicate that AMACR will significantly boost the probability of PCa diagnosis, with post-test values reaching 71% compared to a pre-test baseline of 20%. This suggests its potential as a confirmatory test; however, results outside the confidence region in the HSROC curve should be investigated further.
- Cumulative heterogeneity in both the sensitivity and specificity of AMACAR is a considerable limitation of our meta-analysis (Fig. 3). This variability could be due to clinical, methodological, or statistical issues. From a clinical perspective, the difference in the number of samples analyzed, ranging from 20 by Kaic et al. [30] to 612 by Stewart et al. [25], could potentially have resulted in biased estimates.
- The larger the sample, the higher the likelihood of effect sizes. From a methodological perspective, the differences in populations across 14 countries and five continents may have contributed to the high observed heterogeneity. From a statistical perspective, variation in the reported outcome data likely contributed to the high heterogeneity. However, despite differences in study design, all studies consistently reported high diagnostic accuracy. This uniformity strengthens the likelihood of a normal distribution of diagnostic accuracy estimates within each study and minimizes concerns about statistical heterogeneity.
- Multiple comparable biomarkers were included in several of these studies. For example, the ETS-related gene (ERG) [34] and human telomerase reverse transcriptase [24] displayed perfect specificity but diminished sensitivity levels at 45.1% and 65.96%, respectively. In contrast, Ki-67 [40] and P510s [47] demonstrated flawless sensitivity, yet their specificities were comparatively lower at 66.67% and 25%, respectively. Annexin II (ANXII) [25] and monocarboxylate transporter 2 (MCT2) [35] had a sensitivity of 73.51% and 65.9%, respectively, and specificity of 97.95% and 93.1%.
- Another diagnostic accuracy investigation of PSA was performed by Merriel et al. [67], who reported a combined sensitivity and specificity of 93% and 20%, respectively, across 19 studies. The higher sensitivity and specificity values of AMACR highlight its greater diagnostic accuracy for PCa detection than other currently used biomarkers. However, while PSA is assayed in serum, AMACR is usually assessed from biopsy samples.
- Upon further evaluation across geographical regions, the highest sensitivity and specificity were observed in Asia, based on six studies, with a sensitivity of 95% (95% CI, 89% to 97%) and specificity of 91% (95% CI, 79% to 97%). This was followed by North America, which included 13 studies, with a sensitivity of 89% (95% CI, 83% to 90%) and specificity of 93% (95% CI, 80% to 98%). In Europe, the sensitivity was 81% (95% CI, 74% to 86%) and the specificity was 90% (95% CI, 64% to 98%). Due to the limited number of studies, analyses for South America and Africa could not be performed. Detailed diagnostic performance metrics are presented in Fig. 5. Previous studies have shown that PSA levels vary by patient ethnicity. Black men typically exhibit higher PSA levels and have a higher incidence of PCa diagnoses than White or Hispanic men [68,69]. Differences in AMACR expression across regions and ethnicities have not been well-studied. Further research is needed to evaluate these findings.
- Basal cell markers
- For pathological diagnosis of core needle biopsies, pathologists usually perform high-molecular-weight cytokeratin (HMWCK) (34b12E) in addition to AMACR, so-called “dual IHC staining,” to increase the sensitivity and specificity of IHC. This is a routine process in many pathology laboratories around the world (especially in developed countries).
- The International Society of Urologic Pathology (ISUP) currently advises the use of basal cell markers such as HMWCK, p63, and AMACR to diagnose prostatic adenocarcinoma [70]. While adenocarcinoma generally lacks expression of basal cell markers, benign conditions such as adenosis, atrophy, or benign glands may show similar basal cell loss. In contrast, HMWCK staining in a non-basal pattern, and abnormal diffuse expression of p63 can be seen in PCa. Additionally, AMACR stains 5% to 21% of benign prostatic glands and up to 18% of adenosis cases, which reduces its specificity for diagnosing adenocarcinoma. Therefore, the latest recommendations from the ISUP advise using a combination of those cocktails to evaluate small foci of atypical glands suspected to be prostatic adenocarcinoma [71].
- Strengths and limitations
- This is the first meta-analysis of the diagnostic accuracy of AMACR. We included all sample data regardless of type (tissue biopsy, urine, peripheral blood, and semen) and examination technique (IHC, RT-PCR, ECL-DS, and CLIA). However, a limitation of our study is that, while most studies we reviewed focused on AMACR analysis of biopsy samples via IHC, various other sample types and analysis methods were used; subgroup analyses of HGPIN or Gleason scoring of these studies could not be performed due to their limited numbers.
- Implications for future research and practice
- Conclusive identification of PCa necessitates pathological validation through prostate biopsy, with AMACR immunohistochemistry staining serving as the prevalent supplementary tool to support diagnostic determinations, particularly in complex cases [5]. Prior research reported higher AMACR protein levels in serum [47,48], urine [45,46,49,50], and semen [51] in PCa cases than control cases, suggesting that analysis of AMACR levels in these sample types may be useful as a non-invasive test for PCa. However, the number of studies that have evaluated AMACR expression in non-invasive samples remains limited, and further investigation is needed. Together, our findings indicate that AMACR facilitates screening and early detection of PCa through its high diagnostic accuracy.
- Conclusions
- Our findings validate the high diagnostic accuracy of AMACR in detecting PCa, with strong sensitivity and specificity, particularly in biopsy samples analyzed via immunohistochemistry. While AMACR demonstrates utility in challenging cases, such as atypical presentations and HGPIN, the current evidence remains insufficient. Consequently, it cannot serve as a standalone marker and requires complementary scoring methods. Further research exploring alternative sample types, including serum, urine, and semen, across diverse regions is essential to advance our understanding of AMACR’s diagnostic performance in varied clinical contexts.
DISCUSSION
Supplementary Information
Ethics Statement
Not applicable.
Availability of Data and Material
All data generated or analyzed during the study are included in this published article (and its supplementary information files).
Code Availability
Not applicable.
Author Contributions
Conceptualization: JCP, SKL, NDW. Data curation: SKL, NDW, SA, FXR, AFI. Formal analysis: SKL, NDW. Investigation: JCP, SKL. Methodology: JCP, SKL, NDW, SA, FXR, AFI. Project administration: SKL, NDW. Resources: SKL. Software: SKL, NDW. Supervision: JCP. Validation: JCP. Visualization: SKL, NDW. Writing – original draft: JCP, SKL. Writing – review & editing: JCP, SKL, NDW, SA, FXR, AFI. Approval of final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
No funding to declare.
Acknowledgments
The authors extend their appreciation to Dr Renaningtyas Tambun, Pathologist of St. Carolus Hospital, Jakarta. Dr Johannes Cansius Prihadi is the head of the Urology Department, Faculty of Medicine, Atma Jaya Catholic University, and all colleagues affiliated with Atma Jaya Catholic University of Indonesia for their support and contributions.
| No. | Study | Country | Continent | Size | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Conflict of interest |
|---|---|---|---|---|---|---|---|---|---|---|
| Biopsy/IHC (n = 4,559, sensitivity 90%, specificity 91%) | ||||||||||
| 1 | Yang (2003) [17] | USA | North America | 30 | 100.00 | 86.67 | 88.24 | 100.00 | 93.33 | NA |
| 2 | Browne (2004) [5] | USA | North America | 77 | 90.91 | 93.94 | 95.24 | 88.57 | 92.21 | Academy |
| 3 | Farinola (2004) [18] | USA | North America | 39 | 69.57 | 87.50 | 88.89 | 66.67 | 76.92 | NA |
| 4 | Zhou (2004) [19] | USA | North America | 226 | 81.86 | 63.64 | 97.78 | 15.22 | 80.97 | NA |
| 5 | Hameed (2005) [20] | USA | North America | 359 | 93.65 | 90.00 | 91.24 | 92.73 | 91.92 | NA |
| 6 | Jiang (2005) [21] | USA | North America | 138 | 95.12 | 100.00 | 100.00 | 93.33 | 97.10 | NA |
| 7 | Kunju (2005) [22] | USA | North America | 210 | 86.05 | 93.55 | 90.24 | 90.63 | 90.48 | Academy |
| 8 | Nassar (2005) [23] | USA | North America | 53 | 89.47 | 100.00 | 100.00 | 78.95 | 92.45 | NA |
| 9 | Puebla-Mora (2006) [24] | Mexico | South America | 63 | 90.24 | 72.73 | 86.05 | 80.00 | 84.13 | NA |
| 10 | Stewart (2007) [25] | Canada | North America | 612 | 85.00 | 100.00 | 100.00 | 85.88 | 92.16 | None |
| 11 | Murphy (2007) [26] | Ireland | Europe | 101 | 91.23 | 59.09 | 74.29 | 83.87 | 77.23 | Academy |
| 12 | Ng (2007) [27] | Singapore | Asia | 247 | 98.23 | 94.03 | 93.28 | 98.44 | 95.95 | Academy |
| 13 | Stewart (2008) [28] | Canada | North America | 62 | 63.33 | 75.00 | 70.37 | 68.57 | 69.35 | NA |
| 14 | Trpkov (2009) [29] | Canada | North America | 177 | 96.77 | 58.49 | 84.51 | 88.57 | 85.31 | None |
| 15 | Kaic (2009) [30] | Croatia | North America | 20 | 75.00 | 100.00 | 100.00 | 50.00 | 80.00 | NA |
| 16 | Kumaresan (2010) [31] | India | Asia | 41 | 92.00 | 87.50 | 92.00 | 87.50 | 90.24 | None |
| 17 | Daoud (2012) [32] | Canada | North America | 30 | 70.00 | 100.00 | 100.00 | 62.50 | 80.00 | None |
| 18 | Yamada (2013) [33] | Japan | Asia | 457 | 86.88 | 64.31 | 56.73 | 90.09 | 72.21 | NA |
| 19 | Shah (2013) [34] | USA | North America | 54 | 94.12 | 33.33 | 96.00 | 25.00 | 90.74 | None |
| 20 | Pertega-Gomes (2013) [35] | Portugal | Europe | 552 | 77.36 | 94.09 | 95.74 | 70.74 | 83.51 | None |
| 21 | Murray (2014) [36] | Chile | South America | 559 | 88.52 | 88.03 | 78.26 | 94.03 | 88.19 | NA |
| 22 | Singh (2014) [52] | India | Asia | 80 | 95.00 | 92.50 | 92.68 | 94.87 | 93.75 | None |
| 23 | Alinezhad (2016) [37] | Finland | Europe | 147 | 80.00 | 64.29 | 41.18 | 91.14 | 68.03 | None |
| 24 | Jain (2017) [38] | India | Asia | 50 | 100.00 | 76.47 | 89.19 | 100.00 | 92.00 | NA |
| 25 | Pierconti (2019) [39] | Italy | Europe | 85 | 85.71 | 100.00 | 100.00 | 90.91 | 94.12 | None |
| 26 | Ayowole (2021) [40] | Nigeria | Africa | 60 | 100.00 | 80.00 | 83.33 | 100.00 | 90.00 | NA |
| 27 | Khatun (2023) [41] | India | Asia | 30 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | None |
| Biopsy/RT-PCR (n = 929, sensitivity 83%, specificity 91%) | ||||||||||
| 1 | Jiang (2004) [42] | USA | North America | 731 | 97.14 | 91.70 | 95.04 | 95.13 | 95.08 | NA |
| 2 | Wang (2006) [43] | USA | North America | 86 | 62.71 | 100.00 | 100.00 | 55.10 | 74.42 | Academy |
| 3 | Schostak (2006) [44] | Germany | Europe | 112 | 64.91 | 85.45 | 82.22 | 70.15 | 75.00 | NA |
| Urine/RT-PCR (n = 111, sensitivity 72%, specificity 69%) | ||||||||||
| 1 | Zielie (2004) [45] | USA | North America | 19 | 70.00 | 100.00 | 100.00 | 75.00 | 84.21 | NA |
| 2 | Ouyang (2009) [46] | USA | North America | 92 | 69.77 | 71.43 | 68.18 | 72.92 | 70.65 | Academy |
| Serum/RT-PCR (n = 159, sensitivity 70%, specificity 39%) | ||||||||||
| 1 | Cardillo (2005) [47] | Italy | Europe | 31 | 81.82 | 45.00 | 45.00 | 81.82 | 58.06 | None |
| 2 | Zehentner (2006) [48] | USA | South America | 128 | 67.31 | 36.84 | 42.17 | 62.22 | 49.22 | None |
| Others (n = 140, sensitivity 85%, specificity 62%) | ||||||||||
| 1 | Rogers (2004)a [49] | USA | North America | 26 | 100.00 | 58.33 | 70.59 | 100.00 | 73.08 | NA |
| 2 | Sroka (2015)b [50] | Poland | Europe | 71 | 75.76 | 65.79 | 65.79 | 75.76 | 70.42 | None |
| 3 | Etheridge (2018)c [51] | USA | South America | 43 | 85.71 | 53.33 | 77.42 | 66.67 | 74.42 | Academy |
PPV, positive predictive value; NPV, negative predictive value; IHC, immunohistochemistry; NA, not available; RT-PCR, reverse transcription polymerase chain reaction; ECL-DS, echo chemiluminescence detection system; CLIA, chemiluminescence immunoassay.
aUrine/ECL-DS;
bUrine/CLIA
cSemen/CLIA; size represents biopsy samples.
| No. | Study | Documented converting cases with positive AMACR | Documented converting cases with negative AMACR |
|---|---|---|---|
| 1 | Browne (2004) [5] | 9 HGPIN cases with positive AMACR converted into PCa. | N/A |
| 2 | Zhou (2004) [19] | 34 of 76 Atypical cases converted into PCa (all had AMACR- positive, and 30/34 showed moderate to strong staining). | 39 of 81 Atypical cases have AMACR-negative. All cases were converted into benign based on negative AMACR stain results. |
| 3 | Kunju (2005) [22] | 17 of 32 Atypical cases convert into PCa (13/17 had positive AMACR). | 5 of 17 Atypical cases were AMACR-negative. No atypical AMACR-negative cases converted to PCa. |
| 4 | Stewart (2008) [28] | 27 of 62 HGPIN cases revealed AMACR-positive reactivity and converted into PCa, with at least 1 AMACR-positive HGPIN gland. | From 415 HGPIN glands for evaluation, AMACR-negative reactivity was 35/62 cases. |
| HGPIN cases with positive AMACR were five times more likely to be diagnosed with PCa compared to HGPIN cases with negative AMACR (95% CI, 1.739 to 15.437). | |||
| 5 | Pertega-Gomes (2013) [35] | 31 of 40 HGPIN cases converted into PCa through positive AMACR. | 9 of 40 AMACR-negative HGPIN |
| 6 | Ayowole (2021) [40] | 75% of HGPIN cases with positive AMACR converted into PCa. | 25% AMACR-negative HGPIN |
| 7 | Rogers (2004) [49] | 1 of 2 Atypical cases converted into PCa (1/2 had positive AMACR). | 0 Atypical cases converted into PCa with negative AMACR. Negative AMACR detection were later diagnosed as benign prostate tissue. |
- 1. Rebello RJ, Oing C, Knudsen KE, et al. Prostate cancer. Nat Rev Dis Primers 2021; 7: 9.ArticlePubMedPDF
- 2. Mottet N, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on prostate cancer: 2020 update. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur Urol 2021; 79: 243-62. ArticlePubMed
- 3. Jiang N, Zhu S, Chen J, Niu Y, Zhou L. A-methylacyl-CoA racemase (AMACR) and prostate-cancer risk: a meta-analysis of 4,385 participants. PLoS One 2013; 8: e74386. ArticlePubMedPMC
- 4. Donovan MJ, Cordon-Cardo C. Predicting high-risk disease using tissue biomarkers. Curr Opin Urol 2013; 23: 245-51. ArticlePubMed
- 5. Browne TJ, Hirsch MS, Brodsky G, Welch WR, Loda MF, Rubin MA. Prospective evaluation of AMACR (P504S) and basal cell markers in the assessment of routine prostate needle biopsy specimens. Hum Pathol 2004; 35: 1462-8. ArticlePubMed
- 6. Woodman TJ, Lloyd MD. Analysis of enzyme reactions using NMR techniques: a case study with alpha-methylacyl-CoA racemase (AMACR). Methods Enzymol 2023; 690: 159-209. ArticlePubMed
- 7. Magi-Galluzzi C. Prostate cancer: diagnostic criteria and role of immunohistochemistry. Mod Pathol 2018; 31: S12-21. ArticlePubMedPDF
- 8. Kimura T. East meets West: ethnic differences in prostate cancer epidemiology between East Asians and Caucasians. Chin J Cancer 2012; 31: 421-9. ArticlePubMedPMC
- 9. Ogenyi SI, Okeke BI, Okeke CO, et al. Utilization of diagnostic efficacy of α-methylacyl CoA racemase (AMACR) and p63 in differential diagnosis of prostate cancer and benign prostatic hyperplasia. J Biomed Investig 2024; 12: 103-14.
- 10. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. PLoS Med 2021; 18: e1003583. ArticlePubMedPMC
- 11. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529-36. ArticlePDF
- 12. Cochrane handbook for systematic reviews of diagnostic test accuracy. Cochrane screening and diagnostic tests [Internet]. London: Cochrane Collaboration, 2025 [cited 2025 Mar 26]. Available from: https://methods.cochrane.org/sdt/handbook-dta-reviews.
- 13. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21: 1539-58. ArticlePubMed
- 14. Harbord R. METANDI: Stata module to perform meta-analysis of diagnostic accuracy. Chestnut Hill: Department of Economics, Boston College, 2008.
- 15. Dwamena B. MIDAS: Stata module for meta-analytical integration of diagnostic test accuracy studies [Internet]. Chestnut Hill: Department of Economics, Boston College, 2009 [cited 2024 Mar 23]. Available from: htps://ideas.repec.org//c/boc/bocode/s456880.html.
- 16. Plana MN, Arevalo-Rodriguez I, Fernandez-Garcia S, et al. Meta-DiSc 2.0: a web application for meta-analysis of diagnostic test accuracy data. BMC Med Res Methodol 2022; 22: 306.ArticlePubMedPMCPDF
- 17. Yang XJ, Tretiakova MS, Sengupta E, Gong C, Jiang Z. Florid basal cell hyperplasia of the prostate: a histological, ultrastructural, and immunohistochemical analysis. Hum Pathol 2003; 34: 462-70. ArticlePubMed
- 18. Farinola MA, Epstein JI. Utility of immunohistochemistry for alpha-methylacyl-CoA racemase in distinguishing atrophic prostate cancer from benign atrophy. Hum Pathol 2004; 35: 1272-8. ArticlePubMed
- 19. Zhou M, Aydin H, Kanane H, Epstein JI. How often does alpha-methylacyl-CoA-racemase contribute to resolving an atypical diagnosis on prostate needle biopsy beyond that provided by basal cell markers? Am J Surg Pathol 2004; 28: 239-43. ArticlePubMed
- 20. Hameed O, Sublett J, Humphrey PA. Immunohistochemical stains for p63 and alpha-methylacyl-CoA racemase, versus a cocktail comprising both, in the diagnosis of prostatic carcinoma: a comparison of the immunohistochemical staining of 430 foci in radical prostatectomy and needle biopsy tissues. Am J Surg Pathol 2005; 29: 579-87. ArticlePubMed
- 21. Jiang Z, Li C, Fischer A, Dresser K, Woda BA. Using an AMACR (P504S)/34betaE12/p63 cocktail for the detection of small focal prostate carcinoma in needle biopsy specimens. Am J Clin Pathol 2005; 123: 231-6. ArticlePubMed
- 22. Kunju LP, Chinnaiyan AM, Shah RB. Comparison of monoclonal antibody (P504S) and polyclonal antibody to alpha methylacyl-CoA racemase (AMACR) in the work-up of prostate cancer. Histopathology 2005; 47: 587-96. ArticlePubMed
- 23. Nassar A, Amin MB, Sexton DG, Cohen C. Utility of alpha-methylacyl coenzyme A racemase (p504s antibody) as a diagnostic immunohistochemical marker for cancer. Appl Immunohistochem Mol Morphol 2005; 13: 252-5. ArticlePubMed
- 24. Puebla-Mora AG, Heras A, Cano-Valdez AM, Dominguez-Malagon H. Human telomerase and alpha-methylacyl-coenzyme A racemase in prostatic carcinoma: a comparative immunohistochemical study. Ann Diagn Pathol 2006; 10: 205-8. ArticlePubMed
- 25. Stewart J, Fleshner N, Cole H, Sweet J. Comparison of annexin II, p63 and alpha-methylacyl-CoA racemase immunoreactivity in prostatic tissue: a tissue microarray study. J Clin Pathol 2007; 60: 773-80. ArticlePubMedPMC
- 26. Murphy AJ, Hughes CA, Lannigan G, Sheils O, O'Leary J, Loftus B. Heterogeneous expression of alpha-methylacyl-CoA racemase in prostatic cancer correlates with Gleason score. Histopathology 2007; 50: 243-51. ArticlePubMed
- 27. Ng VW, Koh M, Tan SY, Tan PH. Is triple immunostaining with 34betaE12, p63, and racemase in prostate cancer advantageous? A tissue microarray study. Am J Clin Pathol 2007; 127: 248-53. ArticlePubMed
- 28. Stewart J, Fleshner N, Cole H, Toi A, Sweet J. Prognostic significance of alpha-methylacyl-coA racemase among men with high grade prostatic intraepithelial neoplasia in prostate biopsies. J Urol 2008; 179: 1751-5. ArticlePubMed
- 29. Trpkov K, Bartczak-McKay J, Yilmaz A. Usefulness of cytokeratin 5/6 and AMACR applied as double sequential immunostains for diagnostic assessment of problematic prostate specimens. Am J Clin Pathol 2009; 132: 211-20. ArticlePubMedPDF
- 30. Kaic G, Tomasovic-Loncaric C. Alpha-methylacyl-CoA racemase (AMACR) in fine-needle aspiration specimens of prostate lesions. Diagn Cytopathol 2009; 37: 803-8. ArticlePubMed
- 31. Kumaresan K, Kakkar N, Verma A, Mandal AK, Singh SK, Joshi K. Diagnostic utility of alpha-methylacyl CoA racemase (P504S) & HMWCK in morphologically difficult prostate cancer. Diagn Pathol 2010; 5: 83.ArticlePubMedPMC
- 32. Daoud NA, Li G, Evans AJ, van der Kwast TH. The value of triple antibody (34betaE12 + p63 + AMACR) cocktail stain in radical prostatectomy specimens with crushed surgical margins. J Clin Pathol 2012; 65: 437-40. ArticlePubMed
- 33. Yamada H, Tsuzuki T, Maeda N, et al. Alpha methylacyl-CoA racemase (AMACR) in prostate adenocarcinomas from Japanese patients: is AMACR a "race"-dependent marker? Prostate 2013; 73: 54-9. ArticlePubMedPDF
- 34. Shah RB, Tadros Y, Brummell B, Zhou M. The diagnostic use of ERG in resolving an "atypical glands suspicious for cancer" diagnosis in prostate biopsies beyond that provided by basal cell and alpha-methylacyl-CoA-racemase markers. Hum Pathol 2013; 44: 786-94. ArticlePubMed
- 35. Pertega-Gomes N, Vizcaino JR, Gouveia C, et al. Monocarboxylate transporter 2 (MCT2) as putative biomarker in prostate cancer. Prostate 2013; 73: 763-9. ArticlePubMed
- 36. Murray NP, Reyes E, Fuentealba C, Jacob O, Orellana N. Extended use of P504S positive primary circulating prostate cell detection to determine the need for initial prostate biopsy in a prostate cancer screening program in Chile. Asian Pac J Cancer Prev 2014; 15: 9335-9. ArticlePubMed
- 37. Alinezhad S, Vaananen RM, Ochoa NT, et al. Global expression of AMACR transcripts predicts risk for prostate cancer: a systematic comparison of AMACR protein and mRNA expression in cancerous and noncancerous prostate. BMC Urol 2016; 16: 10.ArticlePubMedPMC
- 38. Jain D, Gupta S, Marwah N, et al. Evaluation of role of alpha-methyl acyl-coenzyme A racemase/P504S and high molecular weight cytokeratin in diagnosing prostatic lesions. J Cancer Res Ther 2017; 13: 21-5. ArticlePubMed
- 39. Pierconti F, Rossi ED, Martini M, Sacco E, Bassi PF, Larocca LM. 34BetaE12 and alfa-methylacyl coenzyme A racemase (AMACR) antibodies better than p63 antibody distinguish normal and neoplastic glands in prostatic tissue with thermal artifacts. Appl Immunohistochem Mol Morphol 2019; 27: 306-10. ArticlePubMed
- 40. Ayowole OA, Hassan YN, Victor E, Toluwalope WH, Adegoke A. Prognostic value of AMACR and Ki-67 in progression of prostate cancer. J Med Dent Sci Res 2021; 8: 41-51.
- 41. Khatun R, Nag D, Saha I. Evaluation of TURP specimen with special reference to prostatic intraepithelial neoplasia and prostatic cancer using light microscopy and immunohistochemical markers of P63, CK5/6, AMACR (p504s), and PSA. Natl J Phys Pharm Pharmacol 2023; 13: 1985-91. Article
- 42. Jiang Z, Wu CL, Woda BA, et al. Alpha-methylacyl-CoA racemase: a multi-institutional study of a new prostate cancer marker. Histopathology 2004; 45: 218-25. ArticlePubMed
- 43. Wang J, Weng J, Cai Y, Penland R, Liu M, Ittmann M. The prostate-specific G-protein coupled receptors PSGR and PSGR2 are prostate cancer biomarkers that are complementary to alpha-methylacyl-CoA racemase. Prostate 2006; 66: 847-57. ArticlePubMed
- 44. Schostak M, Miller K, Krause H, Schrader M, Kempkensteffen C, Kollermann J. Kinetic fluorescence reverse transcriptase-polymerase chain reaction for alpha-methylacyl CoA racemase distinguishes prostate cancer from benign lesions. Cancer Detect Prev 2006; 30: 449-54. ArticlePubMed
- 45. Zielie PJ, Mobley JA, Ebb RG, Jiang Z, Blute RD, Ho SM. A novel diagnostic test for prostate cancer emerges from the determination of alpha-methylacyl-coenzyme a racemase in prostatic secretions. J Urol 2004; 172: 1130-3. ArticlePubMed
- 46. Ouyang B, Bracken B, Burke B, Chung E, Liang J, Ho SM. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol 2009; 181: 2508-13. ArticlePubMedPMC
- 47. Cardillo MR, Gentile V, Ceccariello A, Giacomelli L, Messinetti S, Di Silverio F. Can p503s, p504s and p510s gene expression in peripheral-blood be useful as a marker of prostatic cancer? BMC Cancer 2005; 5: 111.ArticlePubMedPMCPDF
- 48. Zehentner BK, Secrist H, Zhang X, et al. Detection of alpha-methylacyl-coenzyme-A racemase transcripts in blood and urine samples of prostate cancer patients. Mol Diagn Ther 2006; 10: 397-403. ArticlePubMed
- 49. Rogers CG, Yan G, Zha S, et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme a racemase protein. J Urol 2004; 172: 1501-3. ArticlePubMed
- 50. Sroka WD, Adamowski M, Slupski P, et al. Alpha-methylacyl-CoA racemase and hepsin as urinary prostate cancer markers. Int J Biol Markers 2015; 30: e401-6. ArticlePubMedPDF
- 51. Etheridge T, Straus J, Ritter MA, Jarrard DF, Huang W. Semen AMACR protein as a novel method for detecting prostate cancer. Urol Oncol 2018; 36: 532.ArticlePubMed
- 52. Singh V, Manu V, Malik A, Dutta V, Mani NS, Patrikar S. Diagnostic utility of p63 and alpha-methyl acyl Co A racemase in resolving suspicious foci in prostatic needle biopsy and transurethral resection of prostate specimens. J Cancer Res Ther 2014; 10: 686-92. ArticlePubMed
- 53. Ferdinandusse S, Denis S, Clayton PT, et al. Mutations in the gene encoding peroxisomal alpha-methylacyl-CoA racemase cause adult-onset sensory motor neuropathy. Nat Genet 2000; 24: 188-91. ArticlePubMedPDF
- 54. Thompson SA, Calvin J, Hogg S, Ferdinandusse S, Wanders RJ, Barker RA. Relapsing encephalopathy in a patient with alpha-methylacyl-CoA racemase deficiency. J Neurol Neurosurg Psychiatry 2008; 79: 448-50. ArticlePubMed
- 55. McLean BN, Allen J, Ferdinandusse S, Wanders RJ. A new defect of peroxisomal function involving pristanic acid: a case report. J Neurol Neurosurg Psychiatry 2002; 72: 396-9. ArticlePubMedPMC
- 56. Clarke CE, Alger S, Preece MA, et al. Tremor and deep white matter changes in alpha-methylacyl-CoA racemase deficiency. Neurology 2004; 63: 188-9. ArticlePubMed
- 57. Kong G, Lee H, Tran Q, et al. Corrigendum: current knowledge on the function of alpha-methyl acyl-CoA racemase in human diseases. Front Mol Biosci 2021; 8: 639164.ArticlePubMedPMC
- 58. Lee H, Kim M, Kim SH, et al. Alpha-methylacyl-CoA racemase (AMACR), a potential new biomarker for glioblastoma. Front Oncol 2020; 10: 550673.ArticlePubMedPMC
- 59. Jindal Y, Singh A, Kumar R, et al. Expression of alpha methylacyl CoA racemase (AMACR) in gastric adenocarcinoma and its correlation with Helicobacter pylori infection. J Clin Diagn Res 2016; 10: EC10-2. ArticlePubMedPMC
- 60. Shukla N, Adhya AK, Rath J. Expression of alpha-methylacyl-coenzyme A racemase (AMACR) in colorectal neoplasia. J Clin Diagn Res 2017; 11: EC35-8. ArticlePubMedPMC
- 61. Fadare O, Parkash V, Gwin K, et al. Utility of alpha-methylacyl-coenzyme-A racemase (p504s) immunohistochemistry in distinguishing endometrial clear cell carcinomas from serous and endometrioid carcinomas. Hum Pathol 2013; 44: 2814-21. ArticlePubMedPMC
- 62. Eichelberg C, Minner S, Isbarn H, et al. Prognostic value of alpha-methyl CoA racemase (AMACR) expression in renal cell carcinoma. World J Urol 2013; 31: 847-53. ArticlePubMedPDF
- 63. Yu YP, Tsung A, Liu S, et al. Detection of fusion transcripts in the serum samples of patients with hepatocellular carcinoma. Oncotarget 2019; 10: 3352-60. ArticlePubMedPMC
- 64. Taheri D, Roohani E, Izadpanahi MH, et al. Diagnostic utility of a-methylacyl COA racemase in prostate cancer of the Iranian population. J Res Med Sci 2021; 26: 46.ArticlePubMedPMC
- 65. Ji J, Chen X, Xu Y, et al. Prostate cancer diagnosis using urine sediment analysis-based alpha-methylacyl-CoA racemase score: a single-center experience. Cancer Control 2019; 26: 1073274819887697.ArticlePubMedPMC
- 66. Luo J, Zha S, Gage WR, et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res 2002; 62: 2220-6. PubMed
- 67. Merriel SW, Pocock L, Gilbert E, et al. Systematic review and meta-analysis of the diagnostic accuracy of prostate-specific antigen (PSA) for the detection of prostate cancer in symptomatic patients. BMC Med 2022; 20: 54.ArticlePubMedPMCPDF
- 68. Barlow M, Down L, Mounce LTA, et al. Ethnic differences in prostate-specific antigen levels in men without prostate cancer: a systematic review. Prostate Cancer Prostatic Dis 2023; 26: 249-56. ArticlePubMedPMCPDF
- 69. Down L, Barlow M, Bailey SER, et al. Association between patient ethnicity and prostate cancer diagnosis following a prostate-specific antigen test: a cohort study of 730,000 men in primary care in the UK. BMC Med 2024; 22: 82.ArticlePubMedPMCPDF
- 70. Giannico GA, Arnold SA, Gellert LL, Hameed O. New and emerging diagnostic and prognostic immunohistochemical biomarkers in prostate pathology. Adv Anat Pathol 2017; 24: 35-44. ArticlePubMedPMC
- 71. Epstein JI, Egevad L, Humphrey PA, Montironi R; Members of the IIiDUPG. Best practices recommendations in the application of immunohistochemistry in the prostate: report from the International Society of Urologic Pathology consensus conference. Am J Surg Pathol 2014; 38: e6-19. ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Pathogenesis-Guided Biomarker Assessment: A Shift in Prostate Cancer Diagnostics
Jessica M. Logan, Victoria Malone, John J. O’Leary, Doug A. Brooks
International Journal of Molecular Sciences.2025; 26(24): 11786. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
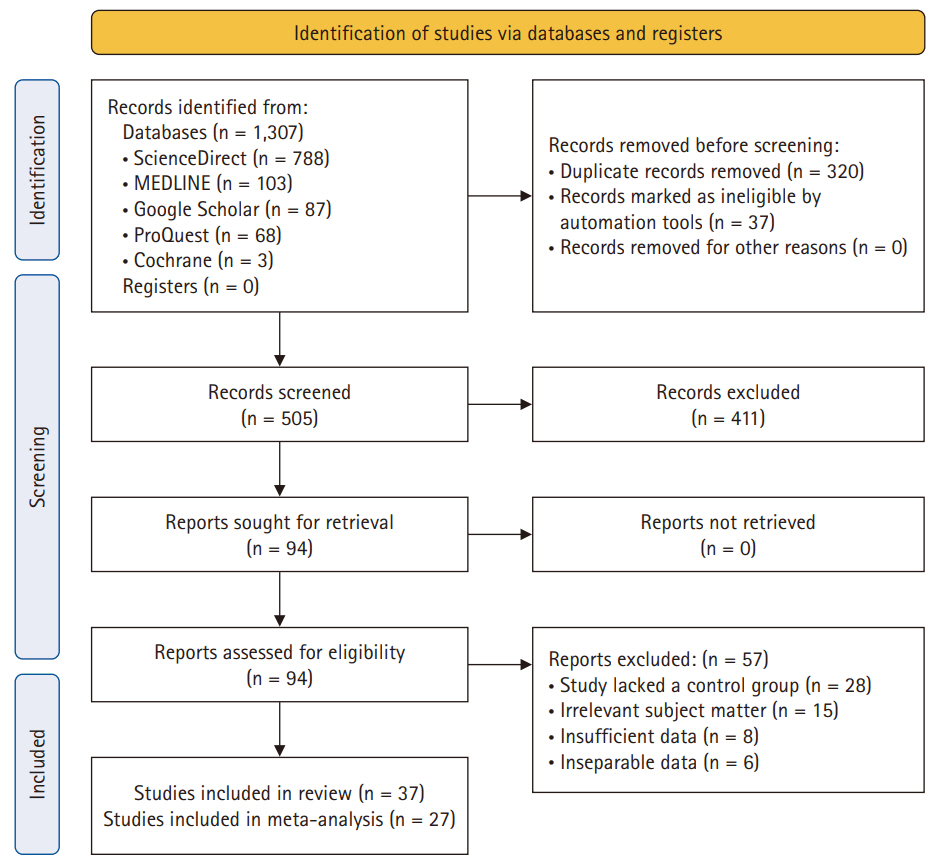
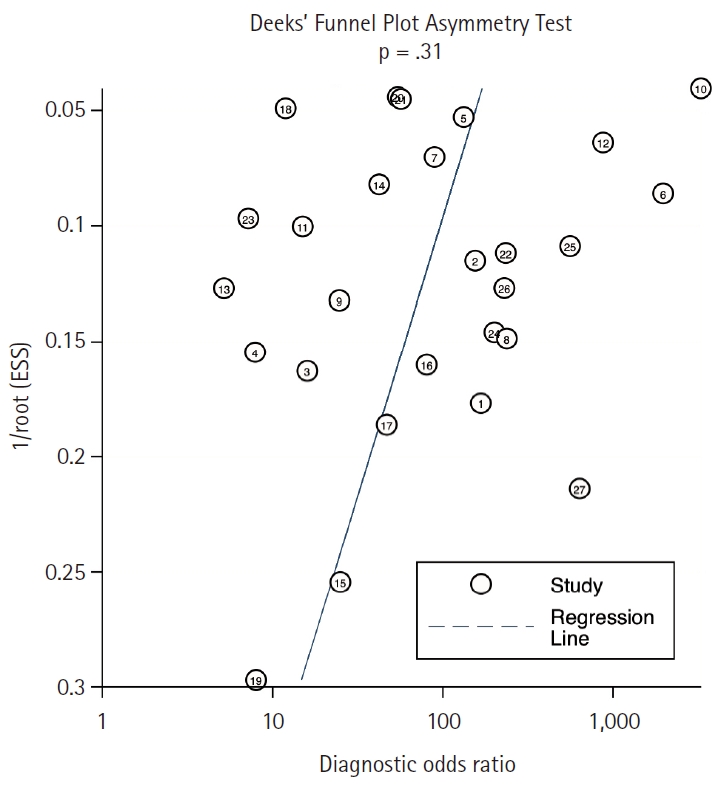
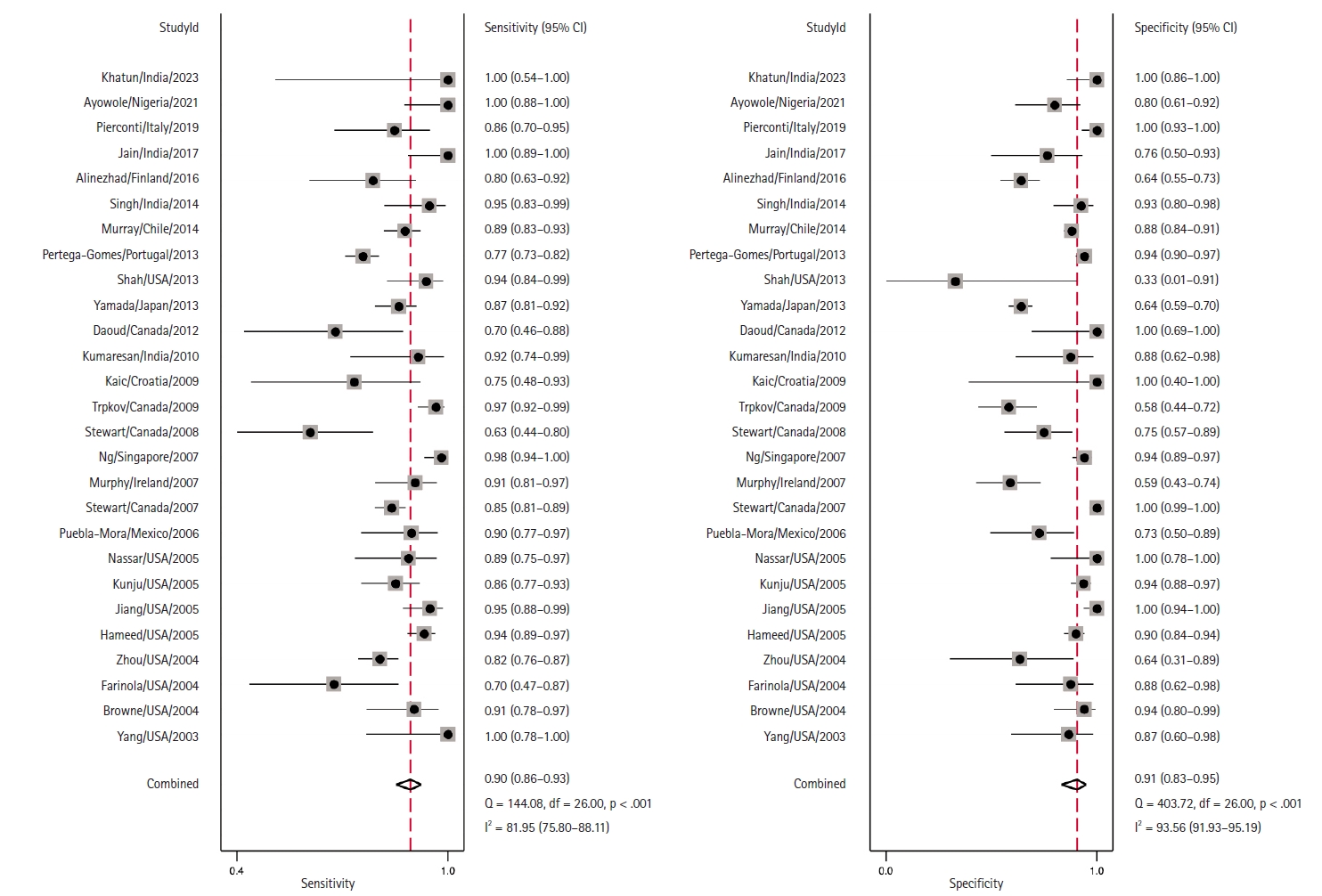
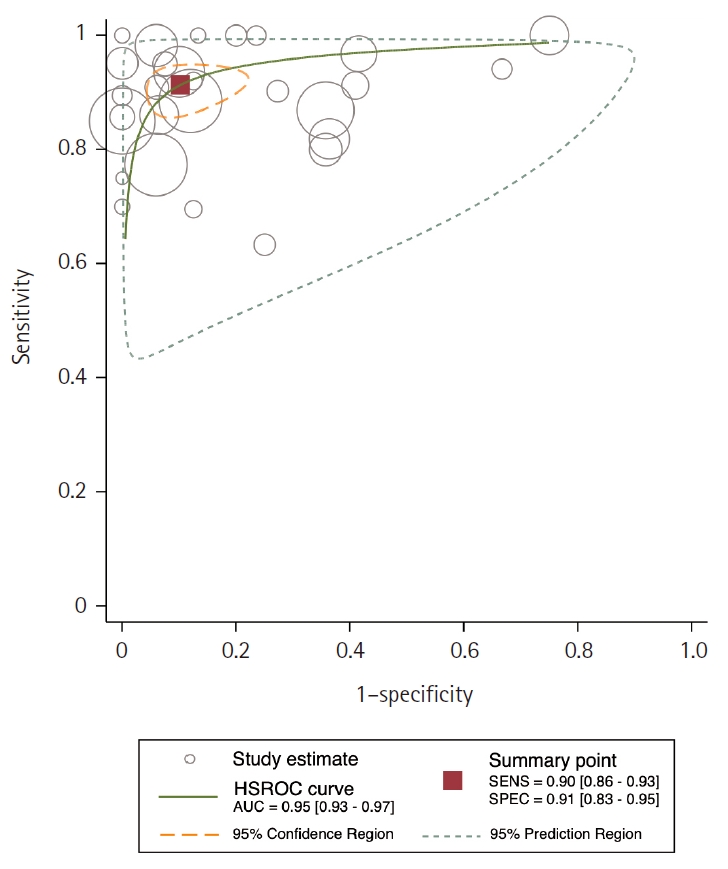
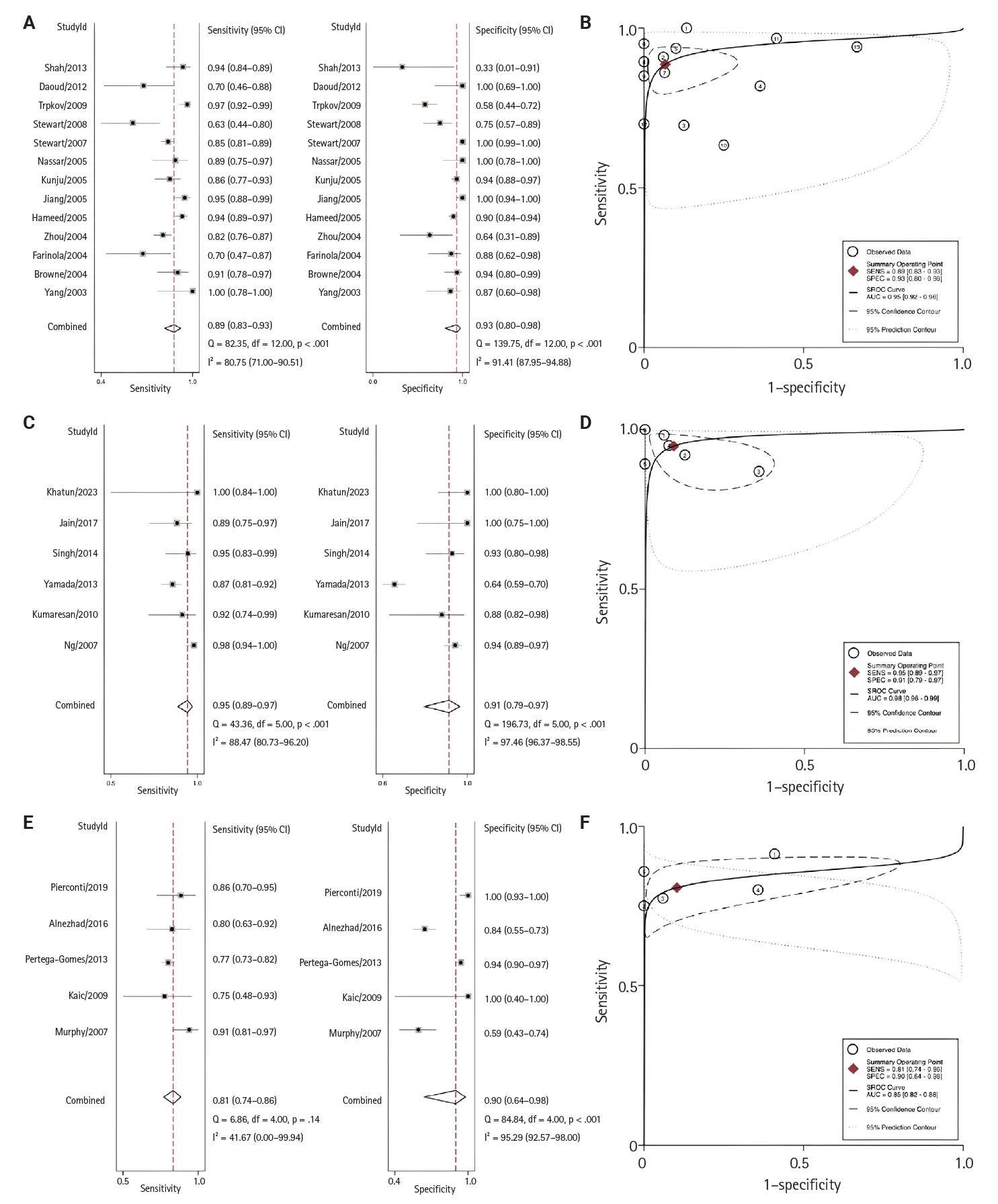
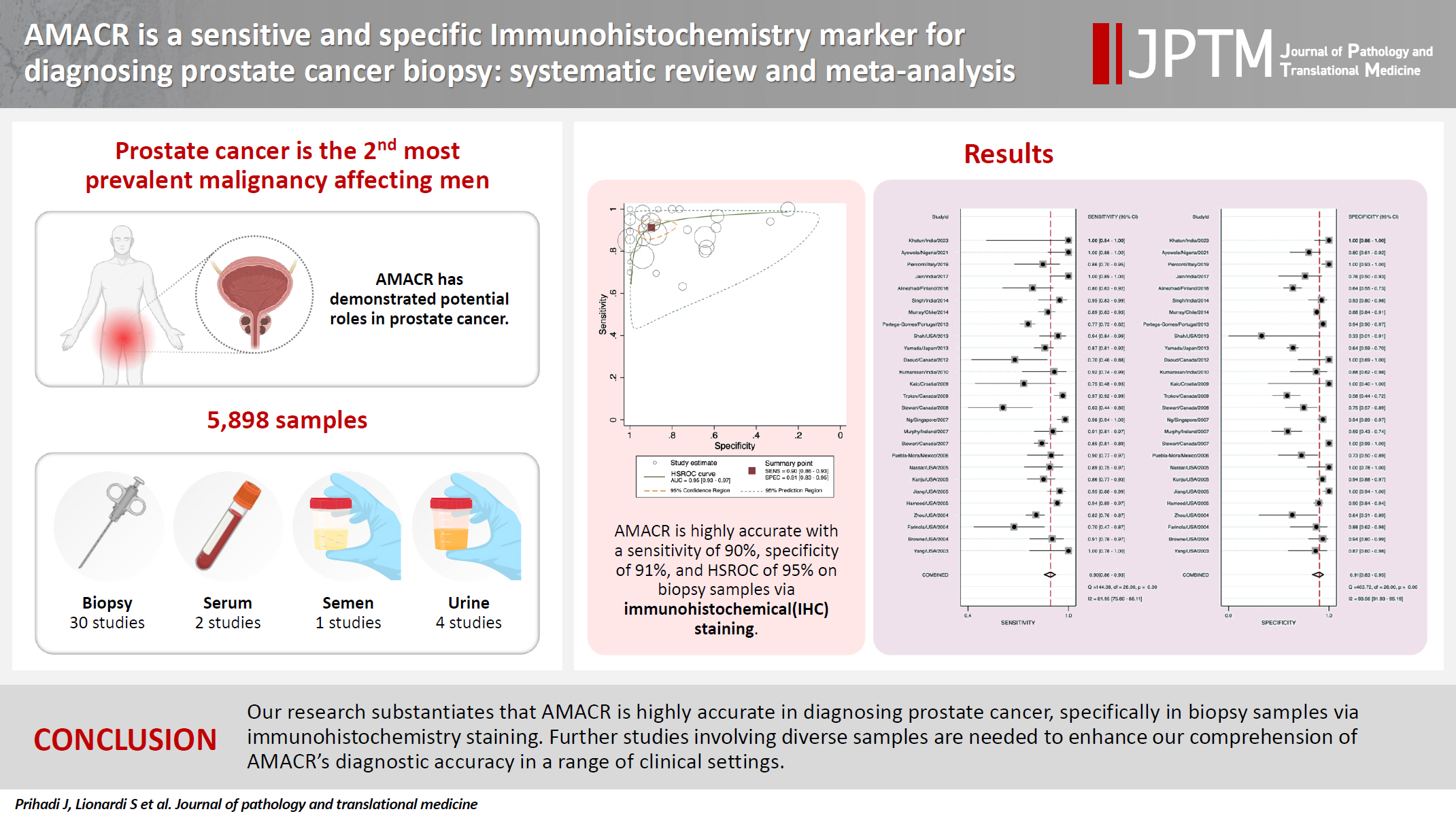
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Fig. 5.
Graphical abstract
| No. | Study | Country | Continent | Size | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Conflict of interest |
|---|---|---|---|---|---|---|---|---|---|---|
| Biopsy/IHC (n = 4,559, sensitivity 90%, specificity 91%) | ||||||||||
| 1 | Yang (2003) [17] | USA | North America | 30 | 100.00 | 86.67 | 88.24 | 100.00 | 93.33 | NA |
| 2 | Browne (2004) [5] | USA | North America | 77 | 90.91 | 93.94 | 95.24 | 88.57 | 92.21 | Academy |
| 3 | Farinola (2004) [18] | USA | North America | 39 | 69.57 | 87.50 | 88.89 | 66.67 | 76.92 | NA |
| 4 | Zhou (2004) [19] | USA | North America | 226 | 81.86 | 63.64 | 97.78 | 15.22 | 80.97 | NA |
| 5 | Hameed (2005) [20] | USA | North America | 359 | 93.65 | 90.00 | 91.24 | 92.73 | 91.92 | NA |
| 6 | Jiang (2005) [21] | USA | North America | 138 | 95.12 | 100.00 | 100.00 | 93.33 | 97.10 | NA |
| 7 | Kunju (2005) [22] | USA | North America | 210 | 86.05 | 93.55 | 90.24 | 90.63 | 90.48 | Academy |
| 8 | Nassar (2005) [23] | USA | North America | 53 | 89.47 | 100.00 | 100.00 | 78.95 | 92.45 | NA |
| 9 | Puebla-Mora (2006) [24] | Mexico | South America | 63 | 90.24 | 72.73 | 86.05 | 80.00 | 84.13 | NA |
| 10 | Stewart (2007) [25] | Canada | North America | 612 | 85.00 | 100.00 | 100.00 | 85.88 | 92.16 | None |
| 11 | Murphy (2007) [26] | Ireland | Europe | 101 | 91.23 | 59.09 | 74.29 | 83.87 | 77.23 | Academy |
| 12 | Ng (2007) [27] | Singapore | Asia | 247 | 98.23 | 94.03 | 93.28 | 98.44 | 95.95 | Academy |
| 13 | Stewart (2008) [28] | Canada | North America | 62 | 63.33 | 75.00 | 70.37 | 68.57 | 69.35 | NA |
| 14 | Trpkov (2009) [29] | Canada | North America | 177 | 96.77 | 58.49 | 84.51 | 88.57 | 85.31 | None |
| 15 | Kaic (2009) [30] | Croatia | North America | 20 | 75.00 | 100.00 | 100.00 | 50.00 | 80.00 | NA |
| 16 | Kumaresan (2010) [31] | India | Asia | 41 | 92.00 | 87.50 | 92.00 | 87.50 | 90.24 | None |
| 17 | Daoud (2012) [32] | Canada | North America | 30 | 70.00 | 100.00 | 100.00 | 62.50 | 80.00 | None |
| 18 | Yamada (2013) [33] | Japan | Asia | 457 | 86.88 | 64.31 | 56.73 | 90.09 | 72.21 | NA |
| 19 | Shah (2013) [34] | USA | North America | 54 | 94.12 | 33.33 | 96.00 | 25.00 | 90.74 | None |
| 20 | Pertega-Gomes (2013) [35] | Portugal | Europe | 552 | 77.36 | 94.09 | 95.74 | 70.74 | 83.51 | None |
| 21 | Murray (2014) [36] | Chile | South America | 559 | 88.52 | 88.03 | 78.26 | 94.03 | 88.19 | NA |
| 22 | Singh (2014) [52] | India | Asia | 80 | 95.00 | 92.50 | 92.68 | 94.87 | 93.75 | None |
| 23 | Alinezhad (2016) [37] | Finland | Europe | 147 | 80.00 | 64.29 | 41.18 | 91.14 | 68.03 | None |
| 24 | Jain (2017) [38] | India | Asia | 50 | 100.00 | 76.47 | 89.19 | 100.00 | 92.00 | NA |
| 25 | Pierconti (2019) [39] | Italy | Europe | 85 | 85.71 | 100.00 | 100.00 | 90.91 | 94.12 | None |
| 26 | Ayowole (2021) [40] | Nigeria | Africa | 60 | 100.00 | 80.00 | 83.33 | 100.00 | 90.00 | NA |
| 27 | Khatun (2023) [41] | India | Asia | 30 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | None |
| Biopsy/RT-PCR (n = 929, sensitivity 83%, specificity 91%) | ||||||||||
| 1 | Jiang (2004) [42] | USA | North America | 731 | 97.14 | 91.70 | 95.04 | 95.13 | 95.08 | NA |
| 2 | Wang (2006) [43] | USA | North America | 86 | 62.71 | 100.00 | 100.00 | 55.10 | 74.42 | Academy |
| 3 | Schostak (2006) [44] | Germany | Europe | 112 | 64.91 | 85.45 | 82.22 | 70.15 | 75.00 | NA |
| Urine/RT-PCR (n = 111, sensitivity 72%, specificity 69%) | ||||||||||
| 1 | Zielie (2004) [45] | USA | North America | 19 | 70.00 | 100.00 | 100.00 | 75.00 | 84.21 | NA |
| 2 | Ouyang (2009) [46] | USA | North America | 92 | 69.77 | 71.43 | 68.18 | 72.92 | 70.65 | Academy |
| Serum/RT-PCR (n = 159, sensitivity 70%, specificity 39%) | ||||||||||
| 1 | Cardillo (2005) [47] | Italy | Europe | 31 | 81.82 | 45.00 | 45.00 | 81.82 | 58.06 | None |
| 2 | Zehentner (2006) [48] | USA | South America | 128 | 67.31 | 36.84 | 42.17 | 62.22 | 49.22 | None |
| Others (n = 140, sensitivity 85%, specificity 62%) | ||||||||||
| 1 | Rogers (2004) |
USA | North America | 26 | 100.00 | 58.33 | 70.59 | 100.00 | 73.08 | NA |
| 2 | Sroka (2015) |
Poland | Europe | 71 | 75.76 | 65.79 | 65.79 | 75.76 | 70.42 | None |
| 3 | Etheridge (2018) |
USA | South America | 43 | 85.71 | 53.33 | 77.42 | 66.67 | 74.42 | Academy |
| No. | Study | Documented converting cases with positive AMACR | Documented converting cases with negative AMACR |
|---|---|---|---|
| 1 | Browne (2004) [5] | 9 HGPIN cases with positive AMACR converted into PCa. | N/A |
| 2 | Zhou (2004) [19] | 34 of 76 Atypical cases converted into PCa (all had AMACR- positive, and 30/34 showed moderate to strong staining). | 39 of 81 Atypical cases have AMACR-negative. All cases were converted into benign based on negative AMACR stain results. |
| 3 | Kunju (2005) [22] | 17 of 32 Atypical cases convert into PCa (13/17 had positive AMACR). | 5 of 17 Atypical cases were AMACR-negative. No atypical AMACR-negative cases converted to PCa. |
| 4 | Stewart (2008) [28] | 27 of 62 HGPIN cases revealed AMACR-positive reactivity and converted into PCa, with at least 1 AMACR-positive HGPIN gland. | From 415 HGPIN glands for evaluation, AMACR-negative reactivity was 35/62 cases. |
| HGPIN cases with positive AMACR were five times more likely to be diagnosed with PCa compared to HGPIN cases with negative AMACR (95% CI, 1.739 to 15.437). | |||
| 5 | Pertega-Gomes (2013) [35] | 31 of 40 HGPIN cases converted into PCa through positive AMACR. | 9 of 40 AMACR-negative HGPIN |
| 6 | Ayowole (2021) [40] | 75% of HGPIN cases with positive AMACR converted into PCa. | 25% AMACR-negative HGPIN |
| 7 | Rogers (2004) [49] | 1 of 2 Atypical cases converted into PCa (1/2 had positive AMACR). | 0 Atypical cases converted into PCa with negative AMACR. Negative AMACR detection were later diagnosed as benign prostate tissue. |
PPV, positive predictive value; NPV, negative predictive value; IHC, immunohistochemistry; NA, not available; RT-PCR, reverse transcription polymerase chain reaction; ECL-DS, echo chemiluminescence detection system; CLIA, chemiluminescence immunoassay. Urine/ECL-DS; Urine/CLIA Semen/CLIA; size represents biopsy samples.
HGPIN, high-grade prostatic intraepithelial neoplasia; AMACR, alpha-methylacyl-CoA racemase; PCa, prostate cancer; N/A, not available; CI, confidence interval.

 E-submission
E-submission