Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 59(2); 2025 > Article
-
Original Article
Categorizing high-grade serous ovarian carcinoma into clinically relevant subgroups using deep learning–based histomic clusters -
Byungsoo Ahn
 , Eunhyang Park
, Eunhyang Park
-
Journal of Pathology and Translational Medicine 2025;59(2):91-104.
DOI: https://doi.org/10.4132/jptm.2024.10.23
Published online: February 18, 2025
Department of Pathology, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- Corresponding Author: Eunhyang Park, MD, PhD Department of Pathology, Severance Hospital, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea Tel: +82-2-2228-1793, Fax: +82-2-362-0860, E-mail: epark54@yuhs.ac
© The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Learning Disabilities in the 21st Century: Integrating Neuroscience, Education, and Technology for Better Outcomes
Syed Mohammed Basheeruddin Asdaq, Ahmad H. Alhowail, Syed Imam Rabbani, Naira Nayeem, Syed Mohammed Emaduddin Asdaq, Faiqa Nausheen
SAGE Open.2025;[Epub] CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure





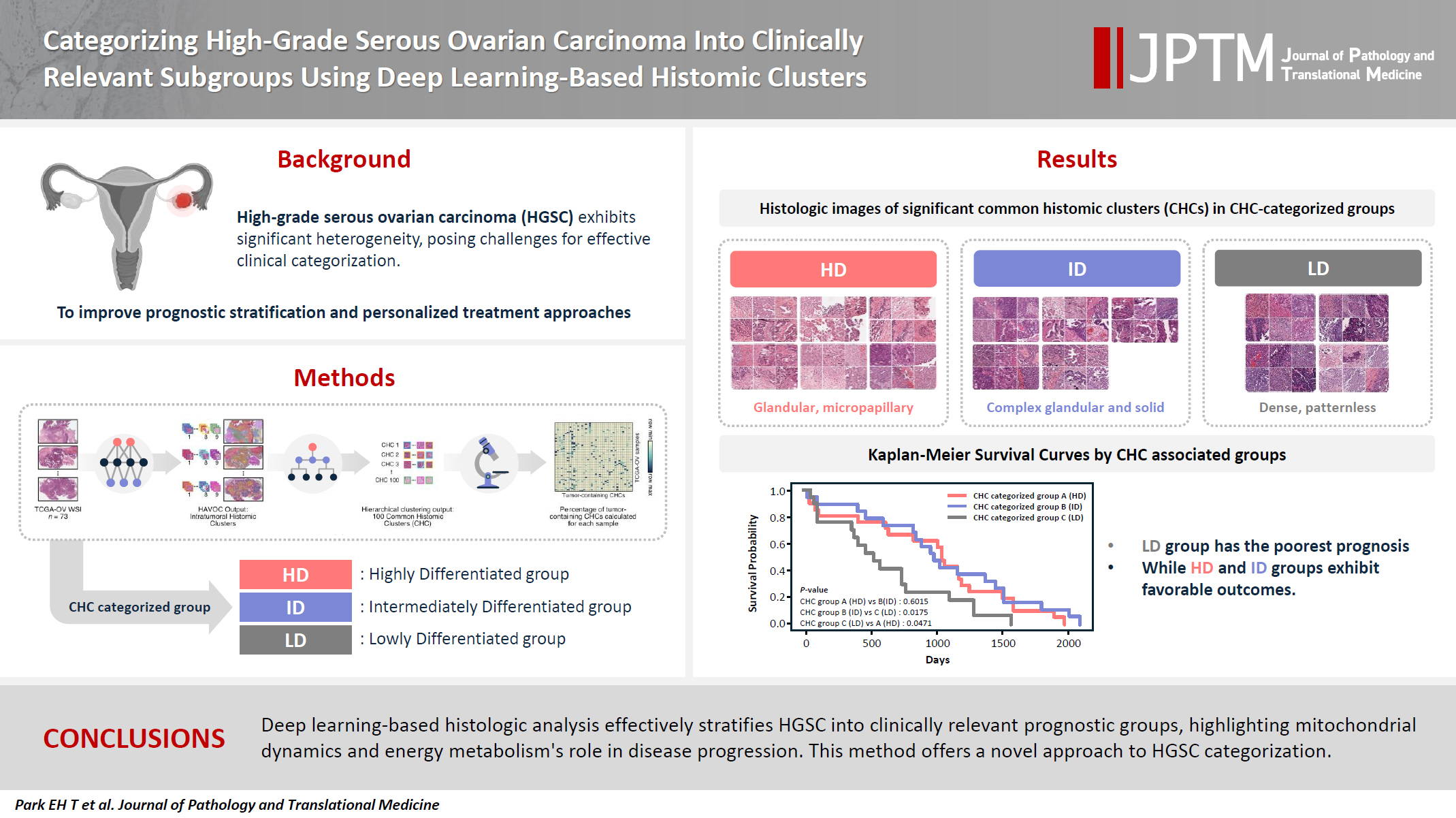
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Fig. 5.
Graphical abstract
| Characteristic | All samples (n = 73) | Group A |
Group B |
Group C |
p-value |
|---|---|---|---|---|---|
| HD group (n = 22) | ID group (n = 27) | LD group (n = 24) | |||
| Age (yr) | .471 | ||||
| <63 | 34 (46.6) | 8 (36.4) | 13 (48.1) | 13 (54.2) | |
| ≥63 | 39 (53.4) | 14 (63.6) | 14 (51.9) | 11 (45.8) | |
| FIGO stage | .651 | ||||
| IIC | 1 (1.4) | 0 | 1 (3.7) | 0 | |
| IIIC | 56 (76.7) | 18 (81.8) | 19 (70.4) | 19 (79.2) | |
| IV | 15 (20.5) | 4 (18.2) | 7 (25.9) | 4 (16.7) | |
| Unknown | 1 (1.4) | 0 | 0 | 1 (4.2) | |
| Molecular classification | .523 | ||||
| Differentiated | 18 (24.7) | 5 (22.7) | 9 (33.3) | 4 (16.7) | |
| Immunoreactive | 10 (13.7) | 4 (18.2) | 3 (11.1) | 3 (12.5) | |
| Mesenchymal | 16 (21.9) | 6 (27.3) | 4 (14.8) | 6 (25.0) | |
| Proliferative | 29 (39.7) | 7 (31.8) | 11 (40.7) | 11 (45.8) | |
| HRD status | .471 | ||||
| HRD | 34 (46.6) | 9 (40.9) | 15 (55.6) | 10 (41.7) | |
| Non-HRD | 34 (46.6) | 10 (45.5) | 10 (37.0) | 14 (58.3) | |
| Unknown | 5 (6.8) | 3 (13.6) | 2 (7.4) | 0 |
| Overall survival | Qualitative histologic analysis | Quantitative histologic analysis |
RNA sequencing |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell features | Color features | Mt & Metab | ECM | Dev & Morph | Metal ion response | Immune response | |||
| Highly differentiated (HD) group | Favorable | Low tumor-to-stromal ratio | Low tumor cell density | High eosin with low hematoxylin staining | +++ | + | + | ++ | +++ |
| High variability among CHCs | Low stromal cell density | ||||||||
| High pleomorphism of tumor cells | Significant nuclei size variations among CHCs | ||||||||
| Includes glandular and micropapillary patterns | Irregular tumor nuclei shape with low circularity and solidity across CHCs | ||||||||
| Intermediately differentiated (ID) group | Favorable | Intermediate tumor-to-stromal ratio | Intermediate tumor cell density | Balanced hematoxylin and eosin staining | ++ | +++ | +++ | +++ | ++ |
| Peritumoral cleft-like spaces | Low stromal cell density | ||||||||
| Includes complex glandular to solid patterns | Large, circular, and solid nuclei | ||||||||
| Little nuclei size variations | |||||||||
| Lowly differentiated (LD) group | Poor | High tumor-to-stromal ratio | High tumor cell density | High hematoxylin and low eosin staining | + | +++ | ++/+++ | + | + |
| Complete patternless sheet-like structure | Low stromal cell density | ||||||||
| Peritumoral cleft-like spaces | Large, circular, and solid nuclei | ||||||||
| Little nuclei size variations | |||||||||
Values are presented as number (%). TCGA-OV, The Cancer Genome Atlas dataset for ovarian cancer; HD, highly differentiated; ID, intermediately differentiated; LD, lowly differentiated; FIGO, International Federation of Gynecology and Obstetrics; HRD, homologous recombination deficiency. Chi-squared test of independence was used to compare the three common histomic cluster–categorized groups.
CHC, common histomic cluster; Mt & Metab, mitochondria and metabolism; ECM, extracellular matrix; Dev & Morph, development and morphogenesis. Legend for RNA sequencing symbols: "+" indicates low expression, "++" represents moderate expression, and "+++" signifies high expression levels across the various categories.

 E-submission
E-submission














