Search
- Page Path
- HOME > Search
Original Articles
- Clinicopathological implications of miR-3127 in melanoma
- Truong Phan-Xuan Nguyen, Minh-Khang Le, Chau M. Bui, Vuong Gia Huy
- J Pathol Transl Med. 2025;59(6):371-381. Published online October 16, 2025
- DOI: https://doi.org/10.4132/jptm.2025.07.08
- 3,044 View
- 138 Download
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
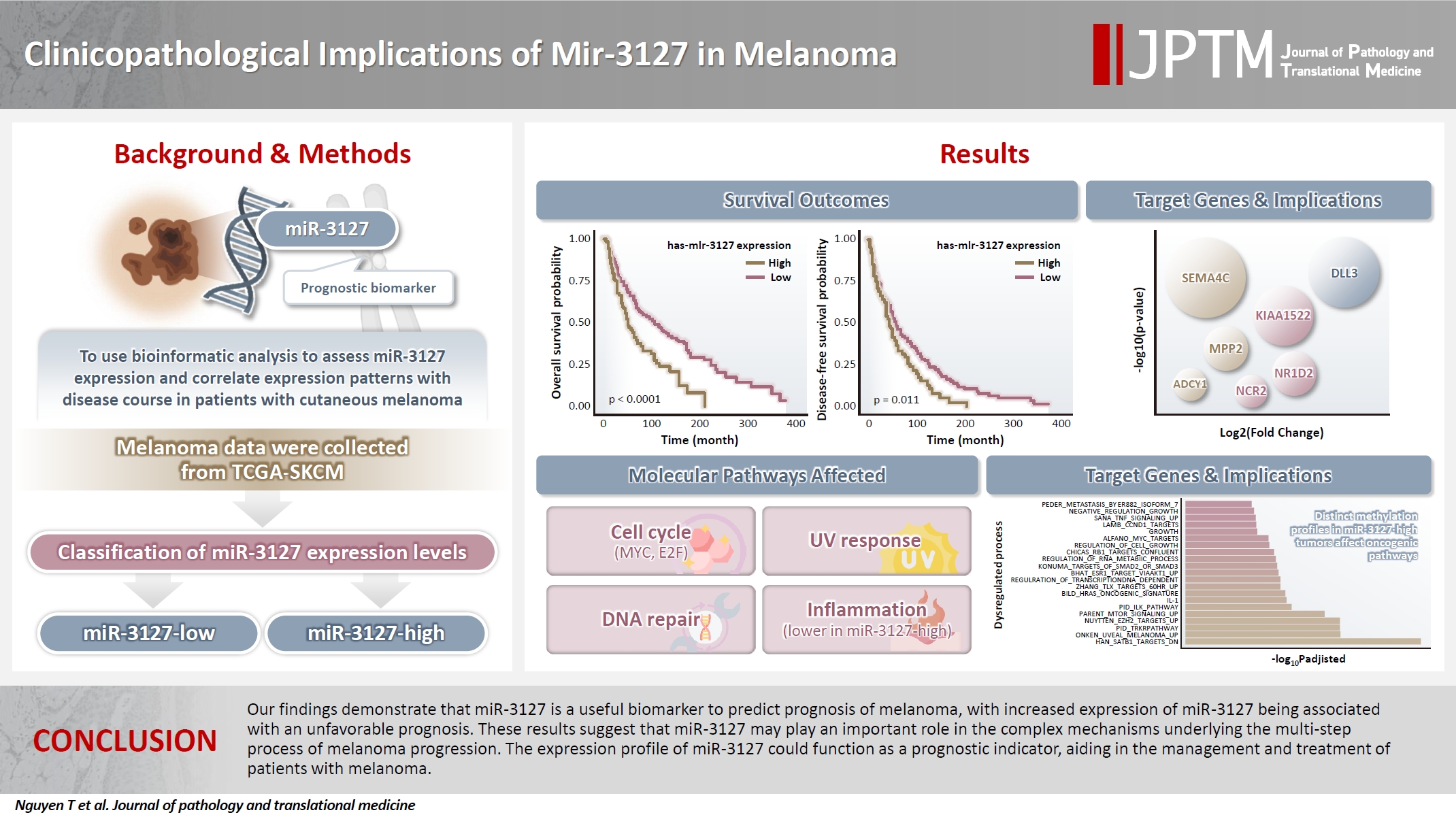
Cutaneous melanoma is the most lethal of all skin cancers. Recent studies suggested that miR-3127 is dysregulated in multiple tumor types and has important roles in tumorigenesis and cancer progression, giving it potential as a prognostic biomarker. The aim of this study was to use bioinformatic analysis to assess miR-3127 expression and correlate expression patterns with disease course in patients with cutaneous melanoma. Methods: miRNA, mRNA sequencing, DNA methylation data, and clinical information of cutaneous melanoma cases were downloaded from the Human Cancer Atlas – Skin Cutaneous Melanoma (TCGA-SKCM). miR-3127 expression was classified into miR-3127–low and miR-3127–high clusters using maximally selected rank statistics. Results: Clustering analysis showed that high expression of miR-3127 (≥20.3 reads per million) was associated with worse progression-free (p < .001) and overall (p = .011) survival compared to low miR-3127 expression. More than five thousand differentially expressed genes between the two miR-3127 sample groups encoded cell differentiation markers, cytokines, growth factors, translocated cancer genes, and oncogenes. Pathway analysis revealed that miR-3127–high samples related to activity of proliferation, DNA repair, and ultraviolet response. Conclusions: The expression level of miR-3127 could act as a prognostic indicator for patients with melanoma.
- Diagnosis of invasive encapsulated follicular variant papillary thyroid carcinoma by protein-based machine learning
- Truong Phan-Xuan Nguyen, Minh-Khang Le, Sittiruk Roytrakul, Shanop Shuangshoti, Nakarin Kitkumthorn, Somboon Keelawat
- J Pathol Transl Med. 2025;59(1):39-49. Published online October 24, 2024
- DOI: https://doi.org/10.4132/jptm.2024.09.14
- 4,158 View
- 324 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
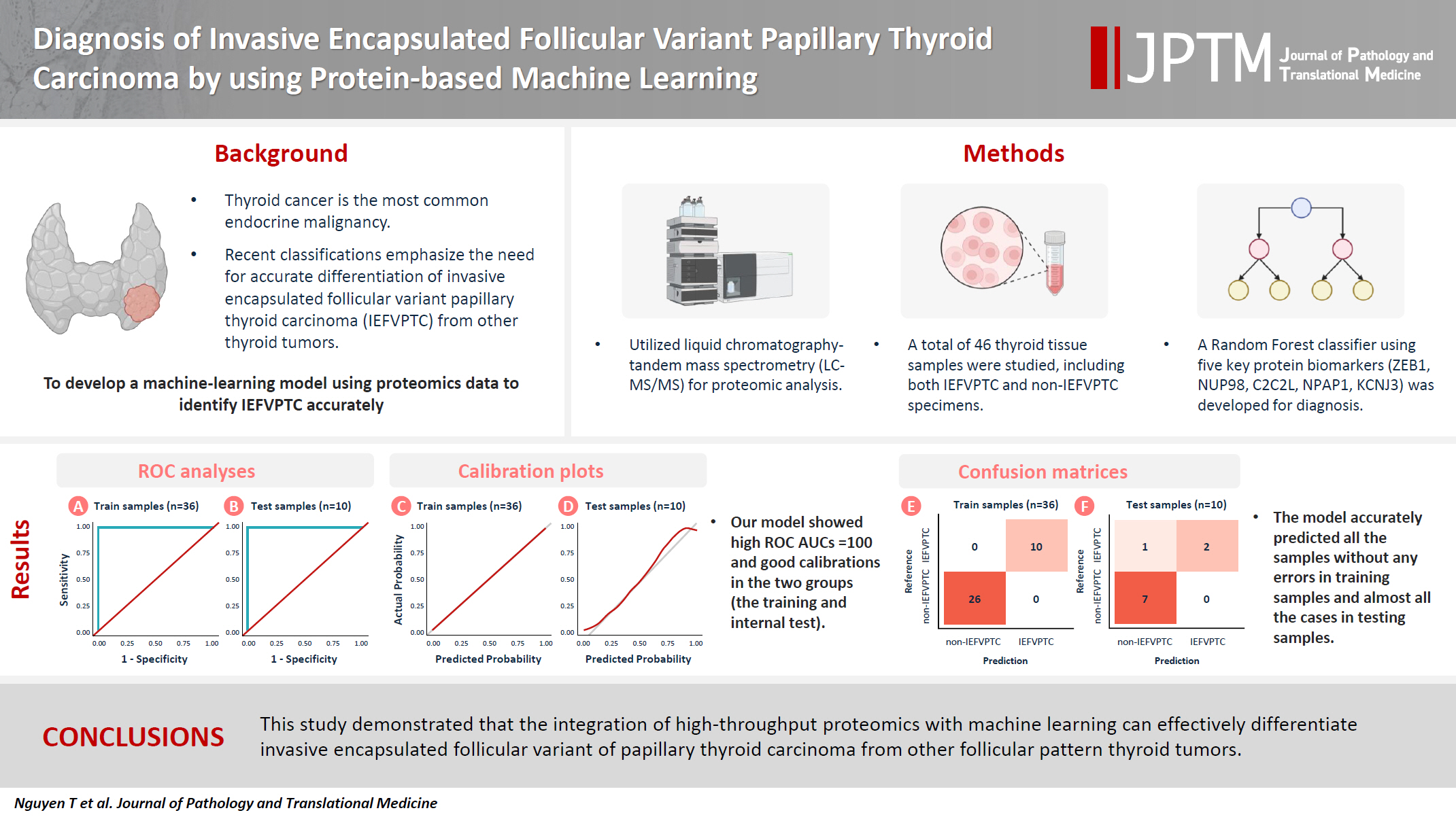
Although the criteria for follicular-pattern thyroid tumors are well-established, diagnosing these lesions remains challenging in some cases. In the recent World Health Organization Classification of Endocrine and Neuroendocrine Tumors (5th edition), the invasive encapsulated follicular variant of papillary thyroid carcinoma was reclassified as its own entity. It is crucial to differentiate this variant of papillary thyroid carcinoma from low-risk follicular pattern tumors due to their shared morphological characteristics. Proteomics holds significant promise for detecting and quantifying protein biomarkers. We investigated the potential value of a protein biomarker panel defined by machine learning for identifying the invasive encapsulated follicular variant of papillary thyroid carcinoma, initially using formalin- fixed paraffin-embedded samples.
Methods
We developed a supervised machine-learning model and tested its performance using proteomics data from 46 thyroid tissue samples.
Results
We applied a random forest classifier utilizing five protein biomarkers (ZEB1, NUP98, C2C2L, NPAP1, and KCNJ3). This classifier achieved areas under the curve (AUCs) of 1.00 and accuracy rates of 1.00 in training samples for distinguishing the invasive encapsulated follicular variant of papillary thyroid carcinoma from non-malignant samples. Additionally, we analyzed the performance of single-protein/gene receiver operating characteristic in differentiating the invasive encapsulated follicular variant of papillary thyroid carcinoma from others within The Cancer Genome Atlas projects, which yielded an AUC >0.5.
Conclusions
We demonstrated that integration of high-throughput proteomics with machine learning can effectively differentiate the invasive encapsulated follicular variant of papillary thyroid carcinoma from other follicular pattern thyroid tumors. -
Citations
Citations to this article as recorded by- Misdiagnosed follicular adenoma with 11 year postoperative liver and lung metastases a case report and literature review
Kai-Li Yang, Heng-Tong Han, Shou-Hua Li, Xiao-Xiao Li, Ze Yang, Li-Bin Ma, Yong-Xun Zhao
Discover Oncology.2025;[Epub] CrossRef
- Misdiagnosed follicular adenoma with 11 year postoperative liver and lung metastases a case report and literature review

 E-submission
E-submission

 First
First Prev
Prev



