Articles
- Page Path
- HOME > J Pathol Transl Med > Forthcoming articles > Article
-
Newsletter
What's new in molecular genetic pathology 2026: emerging biomarkers for personalized cancer therapies -
Umberto Maccio

-
DOI: https://doi.org/10.4132/jptm.2026.01.03
Published online: January 3, 2026
Department of Pathology and Molecular Pathology, University Hospital of Zurich, Switzerland
- Corresponding Author: Umberto Maccio, MD, MSc, MBA Department of Pathology and Molecular Pathology, University Hospital of Zurich, Switzerland Email: umberto.maccio@usz.ch
- This article has been published jointly, with consent, in both Journal of Pathology and Translational Medicine and PathologyOutlines.com.
© The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 61 Views
- 10 Download
Abstract
- New and emerging biomarkers and current molecular assays for the most prevalent and lethal cancers worldwide—breast, lung, prostate, and colorectal cancer—are described. Notably, HER2-low breast cancer and HER2-mutated non-small cell lung cancer have recently been recognized as targetable entities. In addition, various tissue-based analyses are now available to assess prognosis and the risk of relapse in prostate cancer.
- Hormone receptors (estrogen and progesterone receptor), Ki67, and HER2 (per ASCO/CAP guidelines 2023) are assessed in newly diagnosed and relapsed breast cancer (BC). Emerging biomarkers and novel tumor categories are broadening access to targeted therapies.
- ● HER2-low
- ○ HER2-low in BC is defined as HER2 immunohistochemical score of 1+ or 2+ without evidence of HER2 gene amplification via in situ hybridization.
- ○ Since 2022, the antibody-drug conjugate trastuzumab-deruxtecan has been approved for second line treatment in HER2-low BC.
- ● HER2-ultralow (or Her2 Score 0+)
- ○ HER2-ultralow is an emerging category of BC defined as having faint immunohistochemical (IHC) membranous positivity in <10% of tumor cells (formal score 0 according to ASCO/CAP guidelines 2023).
- ○ HER2-ultralow tumors may respond to trastuzumab-deruxtecan, but additional evidence is needed.
- ● Tumor infiltrating lymphocytes (TILs)
- ○ TILs may be associated with a favorable prognosis in triple-negative BC.
- ○ To maximize inter-observer reproducibility and optimize reporting, the International TILs working group has created a website (Home - International TILS Working Group) offering free training.
- ● Next generation sequencing (NGS)
- ○ NGS can be used to analyze advanced BC under current therapies to identify gene mutations with prognostic significance or potential for targeted treatment (Table 1).
BREAST CANCER
- In locally advanced or metastatic non-small cell lung cancer (NSCLC), comprehensive molecular profiling should include NGS panels covering actionable gene alterations, such as EGFR, KRAS, and BRAF, along with IHC assessment of ALK and PD-L1. An NGS panel that incorporates both DNA and RNA analysis enables the detection of gene mutations as well as gene translocations. Liquid biopsy is a minimally invasive procedure, typically performed on peripheral blood, that analyzes circulating tumor DNA (ctDNA) released by tumor cells, through various techniques (NGS, reverse transcriptase polymerase chain reaction [RT-PCR], etc.) with a very low limit of detection (LoD). It is now routinely used in cases of disease progression during targeted therapy to investigate resistance mutations. To ensure the liquid biopsy is representative, the original driver mutations (e.g., EGFR exon 19 deletion) must be detectable. Resistance mutations, if present (e.g., EGFR p.T790M), usually appear in conjunction with the original mutation. Emerging biomarkers and targetable alterations are detailed below.
- ● HER2-gene activating mutations
- ○ NGS is the recommended test to detect HER2 gene mutations in NSCLC, whereas no consensus on IHC testing exists.
- ○ Trastuzumab-emtansine and trastuzumab-deruxtecan showed response rates in over 60% of HER2-mutant NSCLC patients.
- ● MTAP/p16
- ○ Loss of MTAP (typically co-occurring with p16 loss) can be detected through IHC and is a surrogate marker for loss of chromosome 9p21 (Fig. 1).
- ○ This is associated with poor prognosis and resistance against immunotherapy, with clinical trials evaluating response to PRMT5-inhibitors ongoing.
NON-SMALL CELL LUNG CANCER
- Somatic tumor testing using NGS is performed on patients with metastatic castration-resistant prostate cancer (mCRPC) who have progressed on androgen receptor-directed therapy, irrespective of any prior treatment with docetaxel. The National Comprehensive Cancer Network (NCCN) guidelines recommend testing for homologous recombination repair mutations, microsatellite instability-high (MSI-H), deficient mismatch repair (dMMR) genes, and tumor mutational burden (Table 1).
- Moreover, several gene-expression-based tests have recently become available, enabling more refined risk stratification in localized prostate cancer. These tests are approved by NCCN and may be considered in men with NCCN low- or favorable intermediate-risk prostate cancer when additional risk stratification could influence management decisions, such as the choice between active surveillance and definitive therapy.
- The most used gene-expression-based tests approved by NCCN are listed below.
- ● Oncotype DX Genomic Prostate Score
- ○ Ranging from 0-100, it is designed for men with very low- and low-risk prostate cancer to predict the likelihood of organ-confined disease after surgery and has been validated in multiple cohorts.
- ○ The assay analyzes expression of 12 cancer-related genes and 5 housekeeping genes via RT-PCR, focusing on pathways such as cell proliferation, stromal response, and androgen receptor signaling.
- ● Prolaris Prostate Cancer Prognostic Test
- ○ Provides a score (0–6) to estimate 10-year prostate cancer-specific mortality and risk of biochemical recurrence after prostatectomy; it is applicable in both newly diagnosed and post-prostatectomy patients.
- ○ It measures expression of 31 cell cycle genes via quantitative PCR (qPCR), is NCCN-recommended for a wide range of risk categories in patients with ≥10 years life expectancy, and supports decision-making between active surveillance and definitive treatment.
- ● Decipher Prostate Genomic Score
- ○ Provides a score (0–1.0) to predict 5-year metastasis risk, 10-year prostate cancer-specific mortality, and likelihood of high-grade disease to guide the decision between surveillance and treatment in localized cancer, and informs post-prostatectomy management in high-risk cases.
- ○ The assay analyzes expression of 22 genes via microarray, and is recommended by NCCN for patients with low- to intermediate-risk prostate cancer or adverse post-surgical features.
- ● MDx Genomic Prostate Score
- ○ A 17-gene RT-PCR test analyzes prostate tumor tissue to produce a 0–100 aggressiveness score that predicts risks such as adverse pathology, recurrence, metastasis, and cancer-specific mortality, independent of standard clinical factors.
- ○ Although extensively studied within NCCN risk groups and backed by strong clinical evidence, the assay is not formally validated or recommended in current NCCN guidelines.
PROSTATE CANCER
- NGS testing for detection of driver mutations in colorectal cancer (CRC) is recommended in the metastatic setting. Presence of activating mutations in BRAF, KRAS, or NRAS drives therapy choice in a metastatic setting (Table 1). HER2 amplification typically confers resistance against anti-EGFR therapies but represents a potentially targetable alteration. HER2 status should be assessed by IHC; cases with equivocal IHC results should be further evaluated by in situ hybridization. Of note, pathogenic mutations in the POLE or POLD1 gene define a rare subset of CRC characterized by an ultra-mutated phenotype, similar to that seen in endometrial carcinoma. Ultra-mutated CRCs demonstrate an excellent response to immunotherapy.
- Emerging targetable alterations are outlined below.
- ● Mismatch repair (MMR) proteins
- ○ MMR proteins have been routinely assessed in newly diagnosed CRC.
- ○ MLH1 loss with hypermethylation of MLH1 gene promoter and/or BRAF mutation is typically associated with sporadic MMR-deficient CRC.
- ○ Recently, a subgroup of microsatellite-instable CRC BRAFwt with gene-fusions (RET, ALK, ROS1) have been described.
- ● New targetable genes
- ○ NTRK, ALK and ROS1 fusions represent emerging targetable alterations, but occur in a very small subset of colorectal cancers (generally <2%), most often in RAS/BRAF wild-type, right-sided, and frequently mismatch repair-deficient tumors.; therefore, an NGS panel that also evaluates these gene fusions alongside established biomarkers should be considered.
- ○ RET fusions, while actionable in other malignancies (e.g., lung cancer), are not established as a targetable driver in colorectal cancer based on the current evidence. Existing medical literature does not support routine testing or targeting of RET in colorectal cancer.
- ○ Incorporating these targets into routine NGS profiling could expand therapeutic opportunities beyond traditional markers.
COLORECTAL CANCER
- Established biomarkers used in gastric cancer include HER2, PD-L1 and MMR (Table 1).
- A new biomarker is claudin 18.2.
- ● Claudin-18 is normally expressed in gastric mucosa but is downregulated in gastric cancer, particularly at the invasive front, which correlates with increased proliferation and invasion.
- ● The CLDN18::ARHGAP26 fusion is a known driver aberration in diffuse-type gastric cancer.
- ● Claudin 18.2 has emerged as a therapeutic target, with agents such as zolbetuximab demonstrating clinical benefit in claudin 18.2-positive advanced gastric cancer.
- ● Claudin 18.2 is evaluated on IHC by assessing membranous staining of tumor cells, most commonly using validated monoclonal antibodies such as the VENTANA CLDN18 (43-14A) clone. The evaluation is performed on formalin-fixed, paraffin-embedded tissue sections, and the staining is interpreted by a pathologist according to standardized scoring criteria.
- ● The most widely used scoring algorithm, particularly in the context of gastric and gastroesophageal adenocarcinomas, defines positive claudin 18.2 expression as moderate to strong (2+ or 3+) membranous staining in ≥75% of tumor cells.
GASTRIC CANCER
- Dr. Umberto Maccio has been an author and member of the board of reviewers of PathologyOutlines.com since 2024. He graduated from the Medical School in Turin, Italy, and completed thereafter a residency in Anatomical and Surgical Pathology and fellowship in Molecular Genetics at the University Hospital of Zurich, Switzerland, where he currently serves as an attending pathologist and deputy director of the Autopsy Division.
Meet the Author
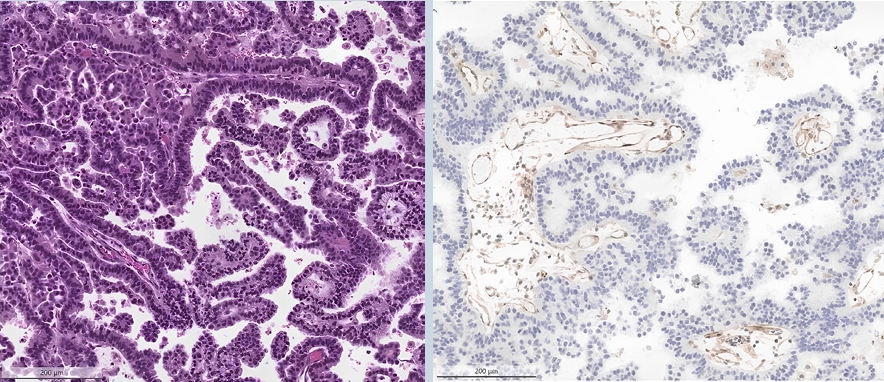
ER: Estrogen receptor; H&E: Hematoxyilin and Eosin; HR: Hormone receptors; HRD: Homologous recombination deficiency; ICI: Immune-checkpoint inhibitor; IHC: Immunohistochemistry; MMR: Mismatch repair; MSI: Microsatellite instability; n/a: not available; NGS: Next generation sequencing; NSCLC: Non-small cell lung carcinoma; PR: Progesterone receptor; TILs: Tumor infiltrating lymphocytes.
Figure & Data
References
Citations

 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
- Related articles
-
- What’s new in adrenal gland pathology: WHO 5th edition for adrenal cortex
- What’s new in bone and soft tissue pathology 2023: guidelines for molecular testing
- What’s new in neuromuscular pathology 2022: myopathy updates and gene therapies
- What’s new in breast pathology 2022: WHO 5th edition and biomarker updates
- What’s new in molecular genetic pathology 2022: immune checkpoint inhibitor biomarkers and select solid tumors
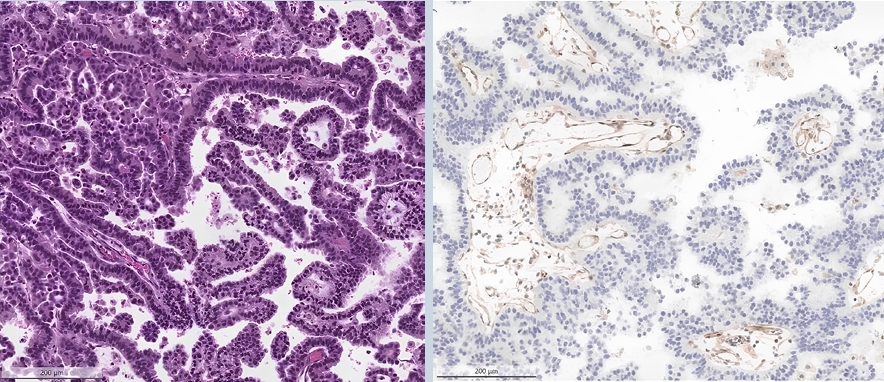
Fig. 1.
| Tumor type | Established Biomarkers/analyses | Clinical Impact/Possible targeted therapies | Recommended testing method | Emerging biomarkers/analyses | Clinical Impact/Possible targeted therapies | Recommended testing method |
|---|---|---|---|---|---|---|
| Breast Cancer | Hormone receptors (ER/PR) | Antihormonal therapies in HR+ BC | IHC | TILs | Better prognosis, response to CT in TNBC | H&E |
| HER2 | HER2-targeted therapies in BC HER2+ and HER2-low | IHC, ISH for HER2 Score 2+ | NTRK-fusions | Targeted therapies in secretory carcinoma (NTRK3::ETV6 translocated) or in other BCs | IHC, NGS | |
| Gene expression test (e.g., OncoTypeDX®, MammaPrint®, EndoPredict®, Prosigna®) | Response to adjuvant CT in HR+ BC | RNA-based tissue analysis | MSI | Therapy with ICIs | IHC (MLH1, PMS2, MSH2, MSH6) or NGS | |
| PIK3CA mutations | Therapy with alpelisib–fulvestrant in HR+ HER2- BC | NGS | AKT1-mutations | Endocrine resistance, Capivasertib in 2nd line | NGS | |
| ESR1 mutations | Resistance against antihormonal therapy Elacestrant in 2nd line | NGS | HRD | Olaparib in BRCA1/2 mutations; possible response to PARP-inhibitors in other HRR-gene mutations (PALB2, CHECK2, etc.) | NGS | |
| Lung cancer (NSCLC) | EGFR mutations | EGFR-targeted therapies | NGS | HER2-gene mutations | HER2-targeted therapies, TKIs | NGS |
| KRAS mutation | Targeted therapy for KRAS p.G12C (sotorasib) | NGS | Tumor mutational burden (TMB) | If high cutoff varying depending on assay) may respond to ICIs | NGS | |
| ALK-translocations | ALK-inhibitors (e.g., alectinib) | IHC, NGS | STK11- and KEAP1-gene mutations | Frequently co-occur: poor survival, may predict resistance against ICIs | NGS | |
| BRAF mutations | BRAF/MEK-inhibitors for BRAF p.V600E | NGS | MTAP and p16 | Loss (surrogate for deletion of chromosome 9p21) associated with poor survival; ongoing trials with PRMT5-inhibitors. | IHC | |
| NTRK-translocations | TKIs (e.g., Entrectinib/Larotrectinib/ | NGS | HRD | Trial with PARP-inhibitors ongoing | NGS | |
| Repotrectinib) | ||||||
| RET-translocations | Selpercatinib | NGS | n/a | |||
| ROS1-translocations | ROS-inhibitors (e.g., crizotinib) | NGS | ||||
| c-MET exon 14 skipping mutations | Tepotinib | NGS | ||||
| PD-L1 | Response to ICIs | IHC | ||||
| Prostate cancer | HRD | PARP-inhibitors | NGS | Oncotype DX Genomic Prostate Score (GPS) | Based on score can help in management/follow-up | |
| MMR | May predict response to ICIs | IHC (MLH1, PMS2, MSH2, MSH6) or NGS or PCR testing Bethesda pentaplex panel | ConfirmMDx | High NPV avoid unnecessary re-biopsy | epigenetic multiplex PCR | |
| TMB | If high (generally >10 mutations/megabase) may predict response to ICIs | NGS | Prolaris test | Based on score can help in management/follow-up | Reverse transcriptase PCR | |
| Androgen Receptor Splice Variant 7 (AR-V7) | Aggressive disease; resistance to abiraterone and enzalutamide; Niclosamide and TAS3681 have been specifically designed to target and inhibit AR-V7, but their clinical use is still being investigated | NGS | Decipher Prostate Genomic Classifier | Based on score can help in management/follow-up | Transcriptomics assay/RNA-based tissue analysis | |
| Colorectal cancer | MMR (MLH1, PMS2, MSH2, MSH6) | If deficient: Rule out Lynch syndrome; response to ICIs; if BRAF wt, screening for targetable gene fusions should be considered. | IHC for MLH1, PMS2, MSH2, MSH6; or NGS or PCR testing Bethesda pentaplex panel | ALK/ROS1/NTRK gene fusions | Potentially targetable | NGS (RNA-based fusion panel); break apart FISH for selected gene translocations |
| BRAF/NRAS/KRAS | Activating mutations predict resistance to anti-EGFR therapy. | NGS | n/a | |||
| KRAS p.G12C can respond to targeted therapy (sotorasib) | ||||||
| HER2 | Amplification confer resistance to anti-EGFR therapy; | IHC for screening; ISH for equivocal cases; NGS to assess mutations | ||||
| Mutations in kinase-domain of ERBB2 may confer resistance to anti-HER2 therapy | ||||||
| POLE-/POLD1-mutations | Usually TMB>100 mut/Mb, demonstrate excellent response to ICIs | NGS | ||||
| Gastric cancer | HER2 | Overexpression or amplification is predictive for response to trastuzumab and guides targeted therapy selection | IHC for screening; ISH for equivocal cases; NGS to assess mutations | Claudin 18.2 | Expression in the tumor cells to select patients eligible for therapy with zolbetuximab | IHC |
| PD-L1 | Expression is used to select patients eligible for therapy with ICIs | IHC, expression assessed through CPS (Combined positive score) | n/a | |||
| MMR (MLH1, PMS2, MSH2, MSH6) | Indicates potential benefit from immunotherapy | IHC | ||||
ER: Estrogen receptor; H&E: Hematoxyilin and Eosin; HR: Hormone receptors; HRD: Homologous recombination deficiency; ICI: Immune-checkpoint inhibitor; IHC: Immunohistochemistry; MMR: Mismatch repair; MSI: Microsatellite instability; n/a: not available; NGS: Next generation sequencing; NSCLC: Non-small cell lung carcinoma; PR: Progesterone receptor; TILs: Tumor infiltrating lymphocytes.

 E-submission
E-submission



