Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 55(2); 2021 > Article
-
Original Article
Deep learning for computer-assisted diagnosis of hereditary diffuse gastric cancer -
Sean A. Rasmussen1
 , Thomas Arnason1
, Thomas Arnason1 , Weei-Yuarn Huang2
, Weei-Yuarn Huang2 -
Journal of Pathology and Translational Medicine 2021;55(2):118-124.
DOI: https://doi.org/10.4132/jptm.2020.12.22
Published online: January 22, 2021
1Department of Pathology and Laboratory Medicine, Queen Elizabeth II Health Sciences Centre and Dalhousie University, Halifax, NS, Canada
2Department of Laboratory Medicine and Molecular Diagnostics, Sunnybrook Health Science Center, University of Toronto, Toronto, ON, Canada
- Corresponding Author: Sean A. Rasmussen, MD, PhD, Division of Anatomical Pathology, Queen Elizabeth II Health Sciences Centre and Dalhousie University, 7th floor, MacKenzie Building, 5788 University Ave, Halifax, NS B3H 1V8, Canada, Tel: +1-289-925-6176, Fax: +1-902-473-1049, E-mail: sean.rasmussen@nshealth.ca
© 2021 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (https://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Patients with hereditary diffuse gastric cancer often undergo prophylactic gastrectomy to minimize cancer risk. Because intramucosal poorly cohesive carcinomas in this setting are typically not grossly visible, many pathologists assess the entire gastrectomy specimen microscopically. With 150 or more slides per case, this is a major time burden for pathologists. This study utilizes deep learning methods to analyze digitized slides and detect regions of carcinoma.
-
Methods
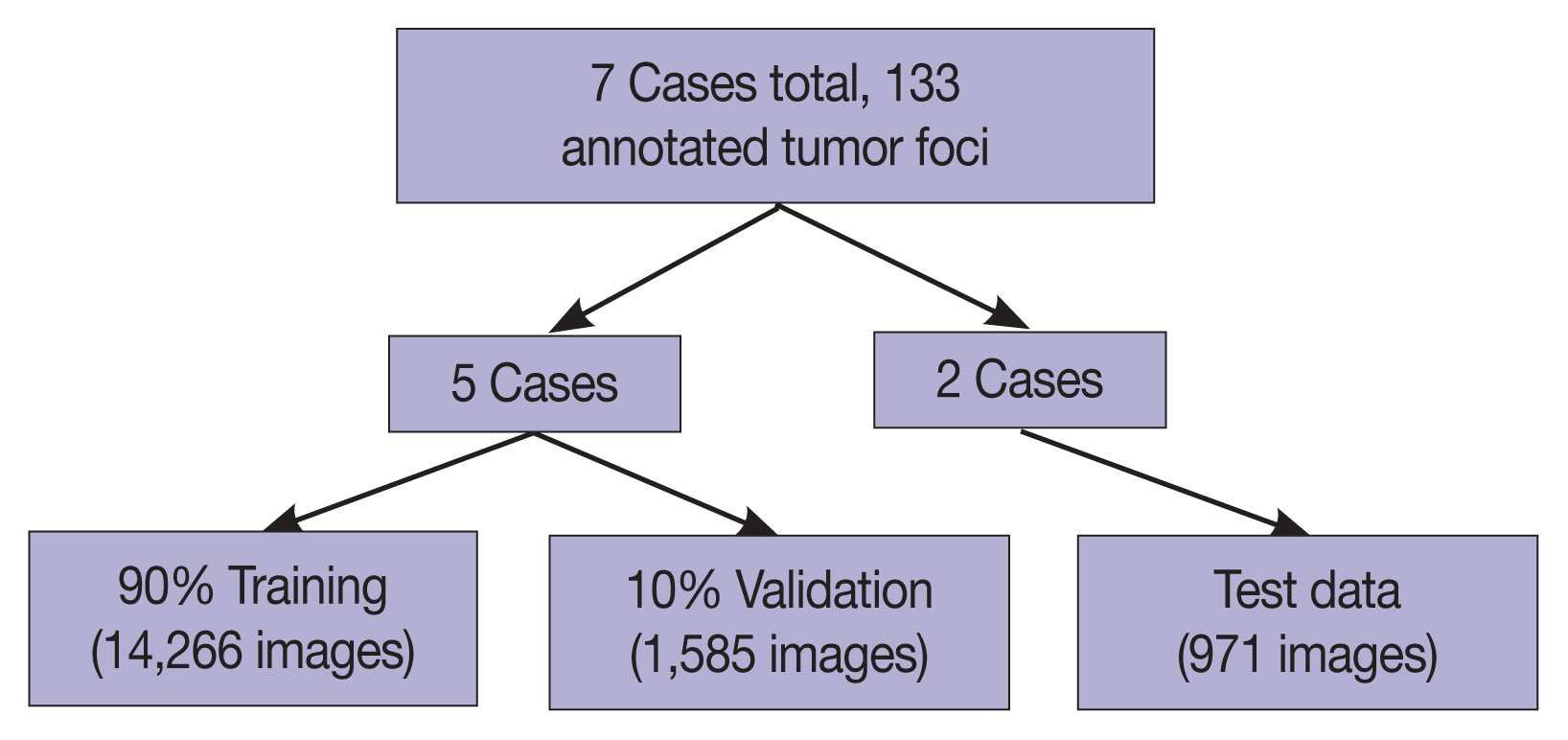
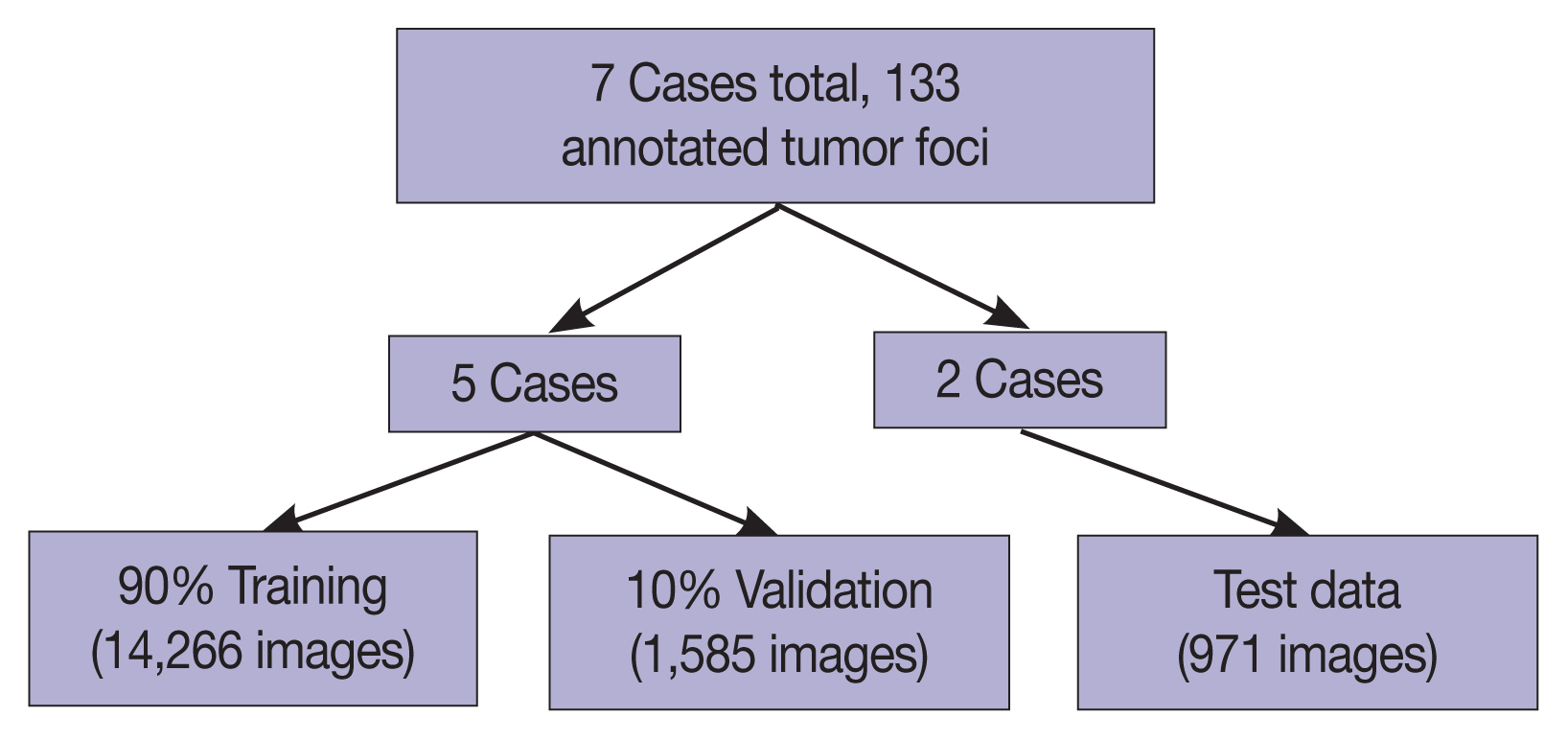
- Prophylactic gastrectomy specimens from seven patients with germline CDH1 mutations were analyzed (five for training/validation and two for testing, with a total of 133 tumor foci). All hematoxylin and eosin slides containing cancer foci were digitally scanned, and patches of size 256 × 256 pixels were randomly extracted from regions of cancer as well as from regions of normal background tissue, resulting in 15,851 images for training/validation and 970 images for testing. A model with DenseNet-169 architecture was trained for 150 epochs, then evaluated on images from the test set. External validation was conducted on 814 images scanned at an outside institution.
-
Results
- On individual patches, the trained model achieved a receiver operating characteristic (ROC) area under the curve (AUC) of 0.9986. This enabled it to maintain a sensitivity of 90% with a false-positive rate of less than 0.1%. On the external validation dataset, the model achieved a similar ROC AUC of 0.9984. On whole slide images, the network detected 100% of tumor foci and correctly eliminated an average of 99.9% of the non-cancer slide area from consideration.
-
Conclusions
- Overall, our model shows encouraging progress towards computer-assisted diagnosis of hereditary diffuse gastric cancer.
- Patients and data collection
- The lab information system at our institution was searched to identify seven consecutive patients with established germline CDH1 mutations who underwent prophylactic total gastrectomy. All patients had foci of intramucosal carcinoma identified microscopically. Patients were not excluded based on age, sex, or medical history. The seven patients we identified included four female and three male patients. The age at the time of gastrectomy ranged from 35 to 59 years (mean, 42.9 years). The number of slides containing intramucosal signet ring cell carcinoma ranged from 5 to 24 per case (mean, 13.4), and the number of lesions per slide ranged from 1 to 7 (mean, 1.4).
- The seven gastrectomy specimens were divided into training and testing datasets. Five cases were used for training and optimizing the model, and two cases were reserved for testing so that no data from these test cases would be seen by the model prior to the final analysis.
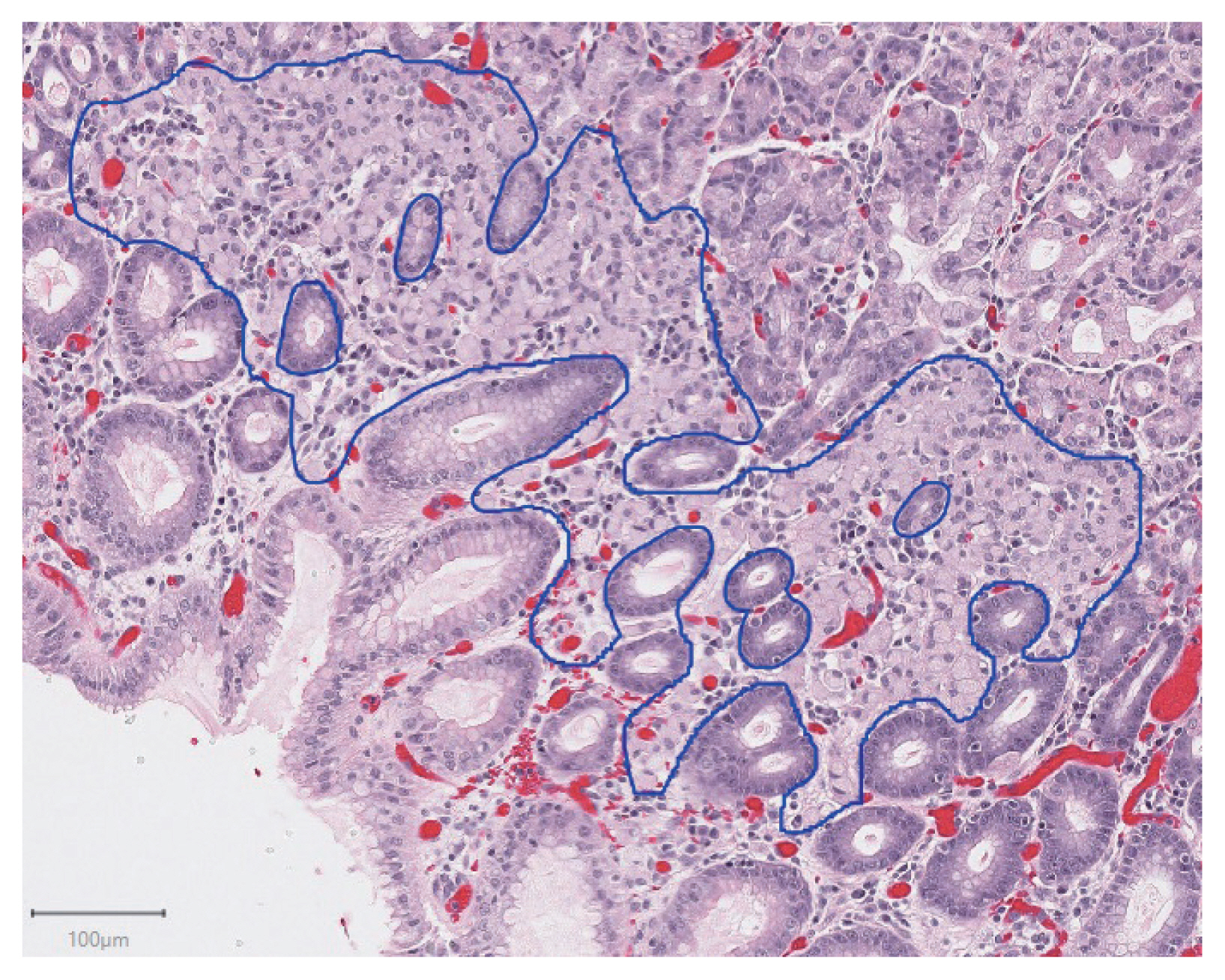
- For all H&E slides from the gastrectomy specimens, foci of intramucosal signet ring cell carcinoma were identified by one of the gastrointestinal pathologists at our institution. The slides with cancer were digitally scanned at 200 × (0.496 μm per pixel) magnification using an Aperio ScanScope XT (Leica Biosystems, Concord, ON, Canada). Within each scanned whole slide image, regions containing cancer were manually annotated by a pathology resident using QuPath v0.2.0-m2 software [11]. Patches of size 256 × 256 pixels were then randomly extracted from these regions such that the central 128 × 128 pixel region overlapped with annotated cancer. The number of patches extracted per lesion was based on the size of the carcinoma focus, such that there was one patch per 2,000 μm2 of carcinoma. In the case of tumor foci smaller than 2,000 μm2, two patches were extracted. To create patches of normal gastric tissue, a total of 150 representative normal regions were digitally scanned (116 from training cases, 34 from test cases). 256 × 256 pixel patches were then randomly extracted from these regions, with the number of extracted patches per region chosen to create roughly balanced datasets in both the training and test groups. For test cases, this resulted in 15 patches per region being extracted, and for training cases (where the tumor foci were larger on average) this resulted in 70 patches per region being extracted.
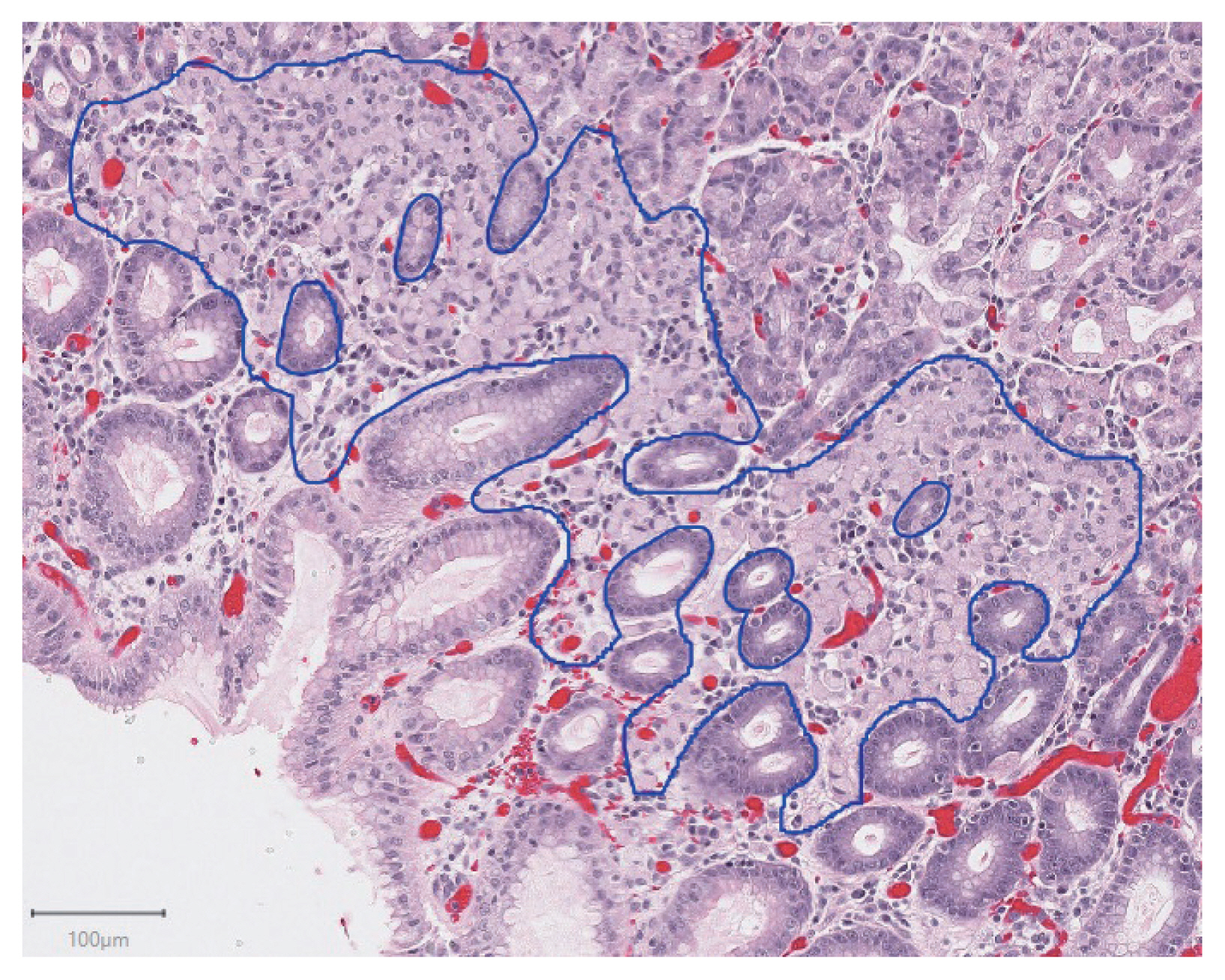
- In total, 94 H&E slides were scanned and 133 individual tumor foci were manually annotated. An example of a manually annotated tumor region is shown in Fig. 1, illustrating the complex borders of some tumor foci. A total of 16,822 patches were extracted, distributed between training and testing sets as shown in Fig. 2. Signet ring cell carcinoma was present in 8,192 of these patches, while the remaining 8,630 patches contained only background gastric tissue.
- To create an external validation dataset, the lab information system at an outside institution (Sunnybrook Health Sciences Centre, Toronto, ON, Canada) was searched to identify recent cases with suspected or confirmed hereditary diffuse gastric cancer. Two cases were identified, with three slides containing carcinoma (out of 110 H&E slides in total). These slides were scanned using the Aperio (Leica Biosystems) scanner at Sunnybrook Health Sciences Centre, and 256 × 256 pixel patches were extracted in a manner identical to that described above for test cases. This resulted in a total of 814 patches, with 394 of these containing carcinoma.
- CNN training
- Image patches from the five training cases were randomly divided into training (90%) and validation (10%) sets. A model was trained using DenseNet-169 architecture with compression of 0.5, dropout of 0.2, and bottleneck layers as described in Huang et al. [8]. The model was coded in Python using TensorFlow v1.14.0 [12] with the Keras API. We trained for 100 epochs with stochastic gradient descent at learning rate 0.1, 30 epochs at learning rate 0.01, and 20 epochs at learning rate 0.001. Momentum for batch normalization was set at 0.99. Data was augmented during training with rotations of 0°, 90°, 180°, or 270°. The model was evaluated on the validation images after each epoch and at the end of training to monitor progress and fine tune training parameters. Various learning rate schedules and minor variations on model architecture were applied. In particular, we found that the addition of dropout layers (which are optional in the DenseNet architecture [8]) was useful to reduce overfitting in this relatively small dataset. Only the most successful model based on validation performance was evaluated on the test set.
- Training was completed on an NVIDIA GeForce GTX 1060 6GB GPU (Nvidia Corporation, Santa Clara, CA, USA).
- Data analysis
- The first issue to address was whether the trained model could accurately classify individual image patches. After training on patches from the first five cases, the model was evaluated on patches extracted from the two test cases. Receiver operating characteristic (ROC) analysis was completed and the area under the curve (AUC) was calculated based on the model’s predicted classification on these images compared to ground truth annotations. An identical analysis was conducted on patches extracted from the external validation cases.
- The second issue to address was whether the trained model could efficiently analyse whole slide images and identify areas suspicious for carcinoma. We used an approach similar to that described by Liu et al. [5]. Whole slide images from the two test cases were tiled into patches of size 256 × 256 pixels with 128 pixels of overlap between adjacent patches. The trained model then made a prediction on each patch. In this way, each tumor focus would be analyzed in multiple overlapping patches, so even if one patch resulted in a false-negative, the tumor focus may still be detected in an adjacent/overlapping patch. When a patch was predicted to be positive, the image was rotated 180° and another prediction was made. The final prediction was called positive only if both individual predictions exceeded the threshold value. The threshold value for classifying a patch as positive for carcinoma was chosen based on the value needed to maintain 90% sensitivity for carcinoma patches in the validation dataset. This approach was chosen in an effort to minimize false-positives, while still maintaining adequate sensitivity for individual tumor foci.
- Sensitivity was calculated for the whole slide image analysis based on the number of tumor foci that overlapped with at least one patch predicted positive by the model. We also calculated the false-positive rate as a percentage of the non-cancer slide area that was predicted positive by the model.
MATERIALS AND METHODS
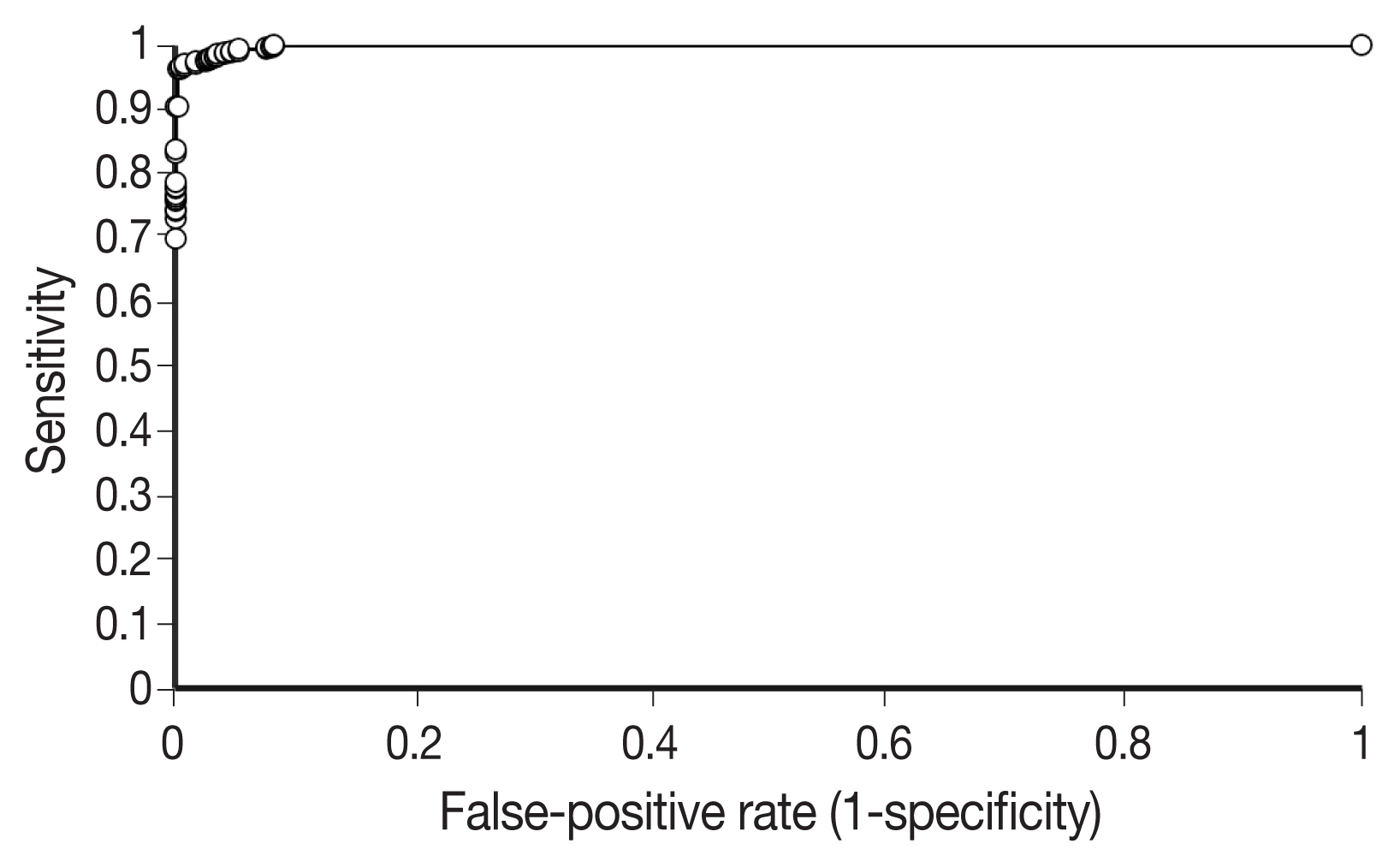
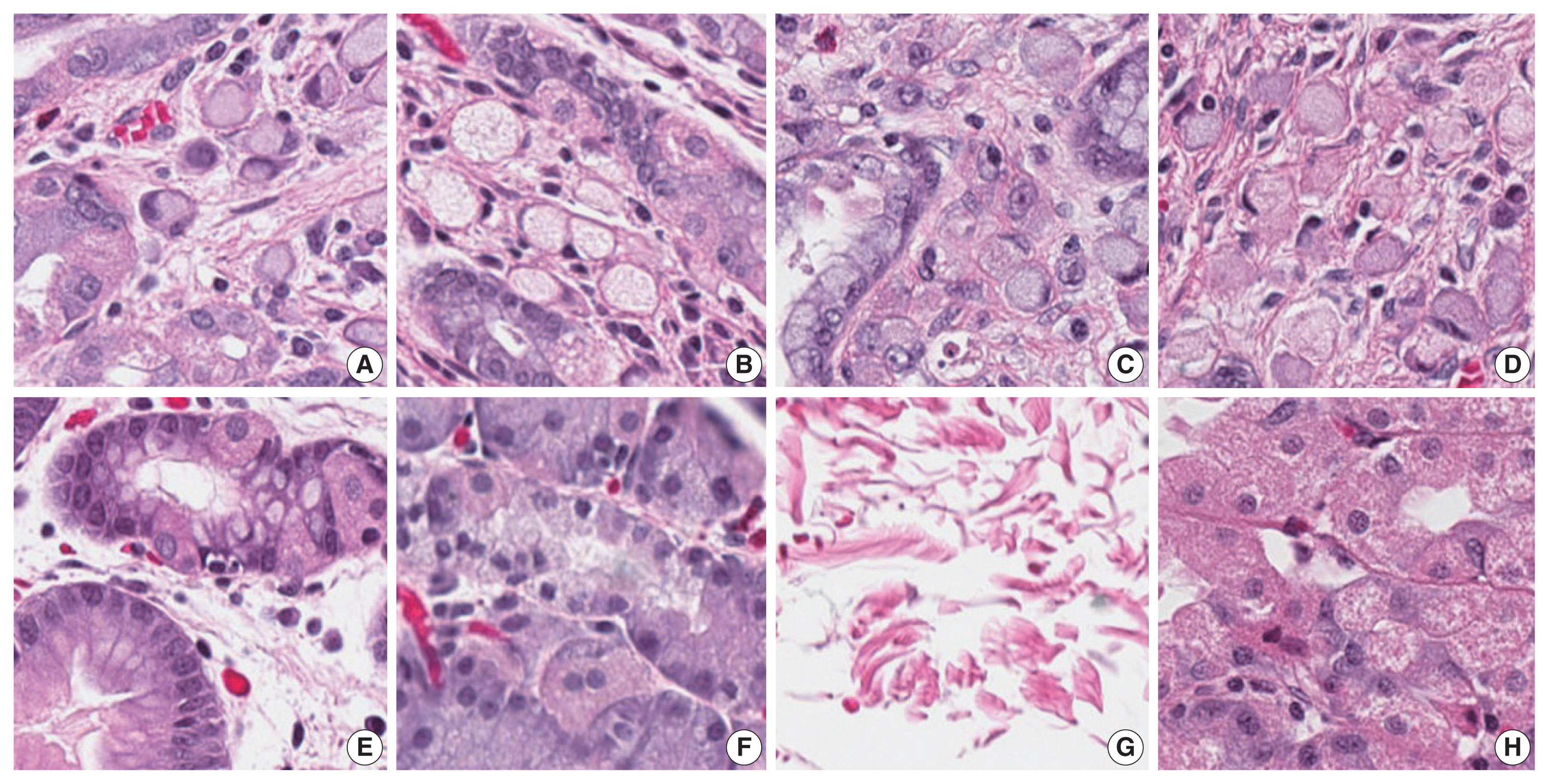
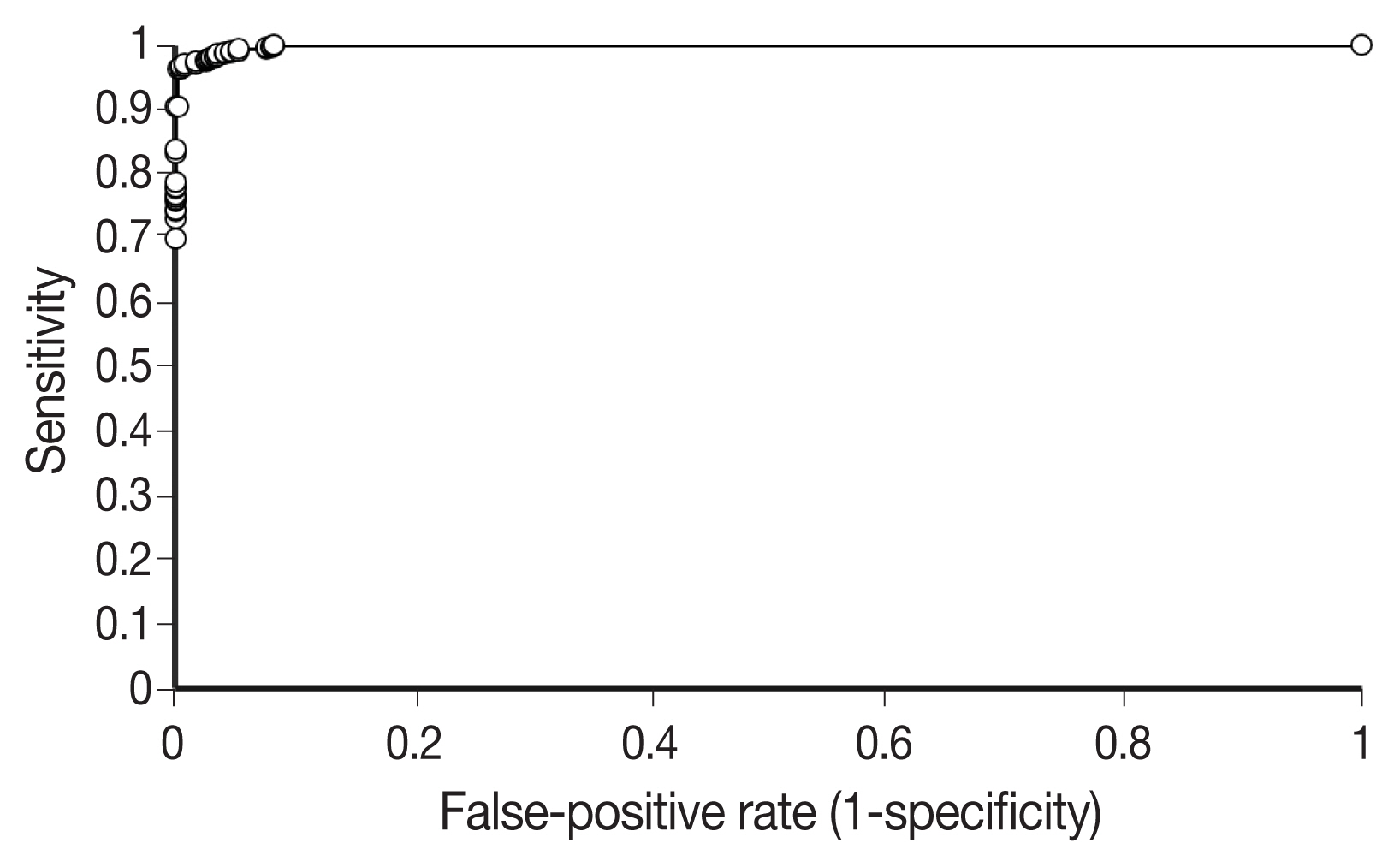
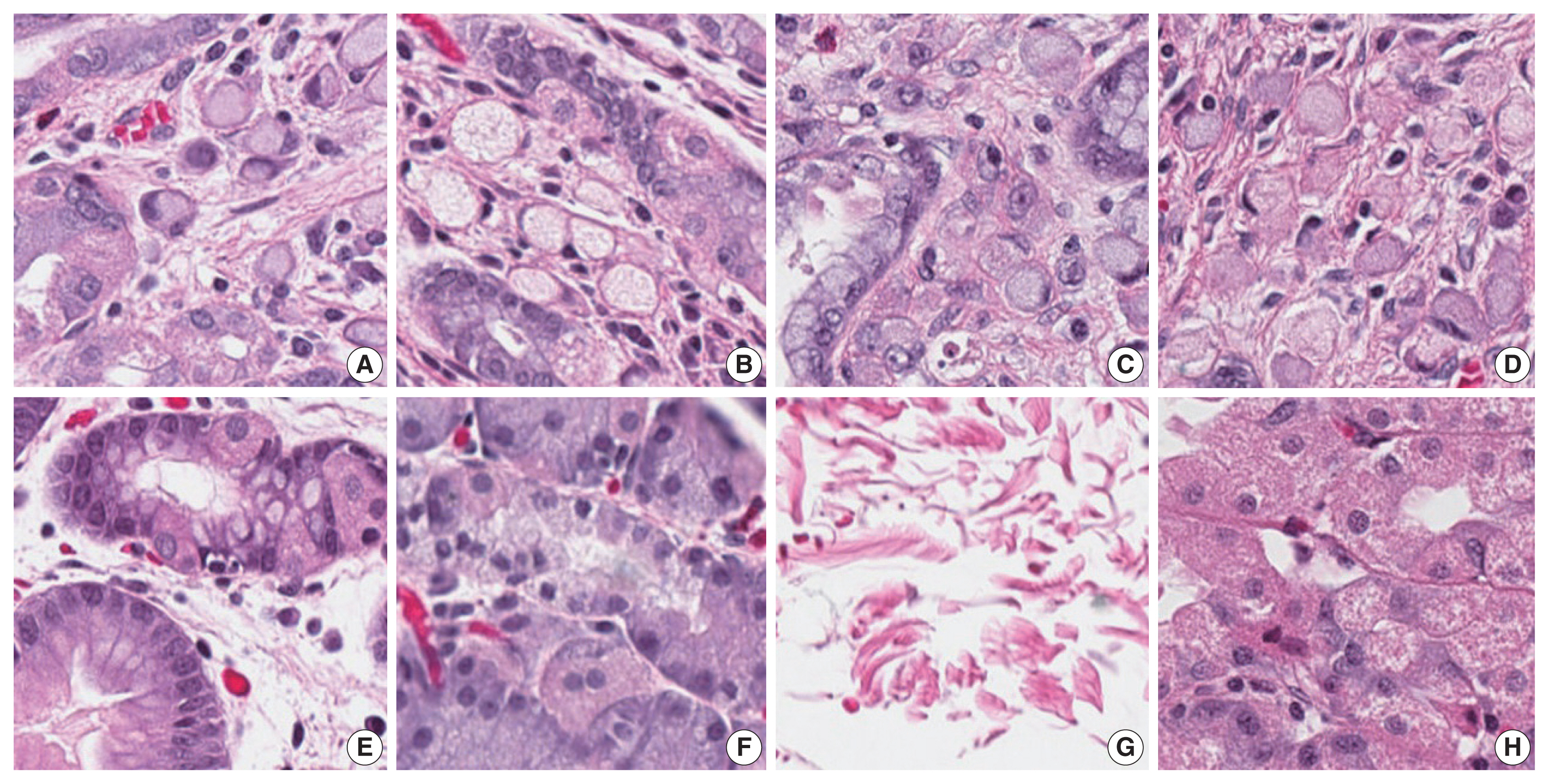
- Our first major objective was to determine whether the trained model could correctly classify individual patches as containing signet ring cell carcinoma or not. To assess this, the trained model was evaluated on patches from the test set, which it had not seen during training. On these images, the trained model achieved an ROC AUC of 0.9986 (Fig. 3). This would permit sensitivity of 95% with a false-positive rate of 0.2%, or a sensitivity of 90% with a false-positive rate of less than 0.1%. Fig. 4 illustrates several examples of correctly classified patches containing signet ring cell carcinoma or normal tissue.
- We conducted an identical analysis on the external validation dataset to determine whether the model’s performance could be generalized to images from slides stained and scanned at an outside institution. On external validation images, the model achieved a similar ROC AUC of 0.9984. This would permit sensitivity of 95% with a false-positive rate of 0.5%, or a sensitivity of 90% with a false-positive rate of less than 0.1%.
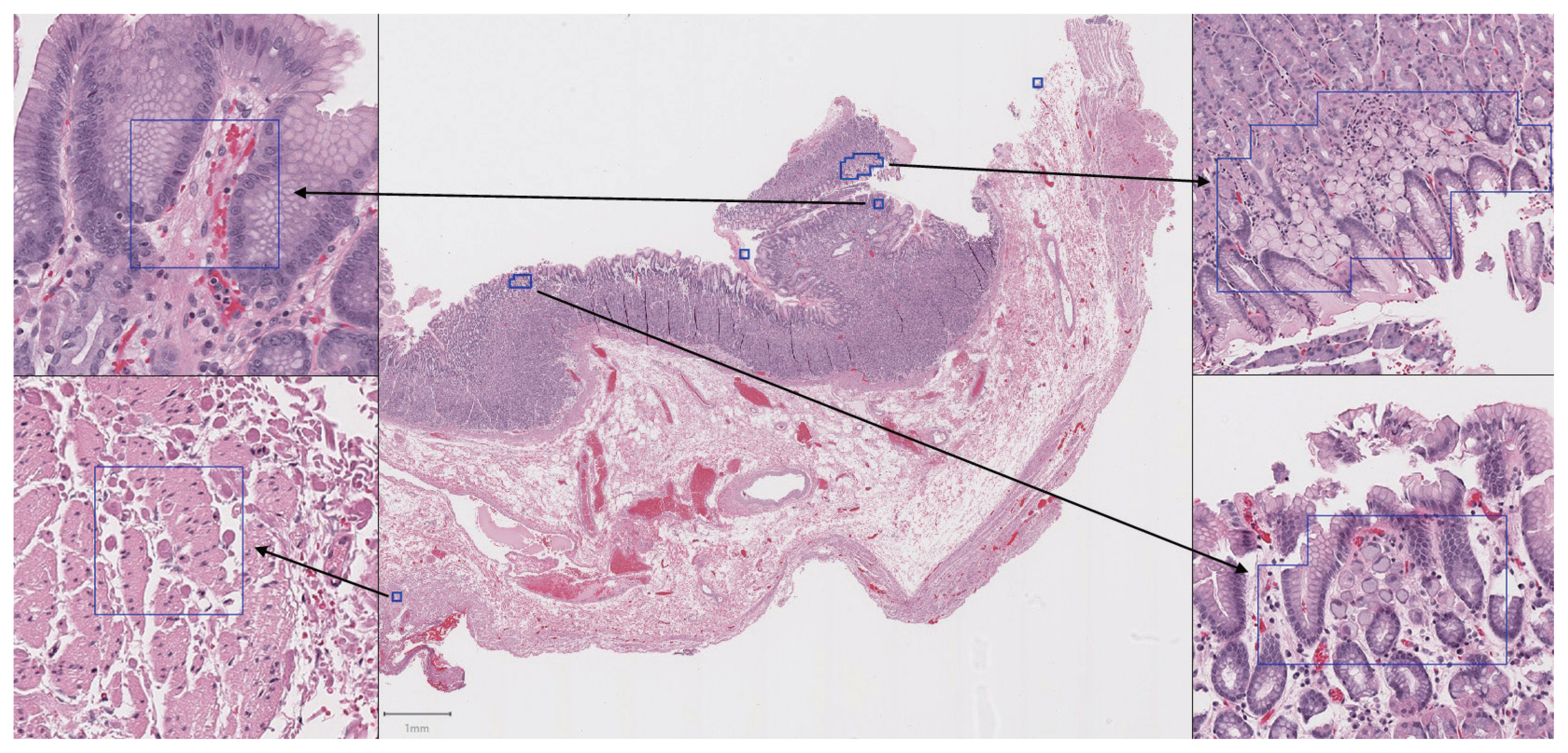
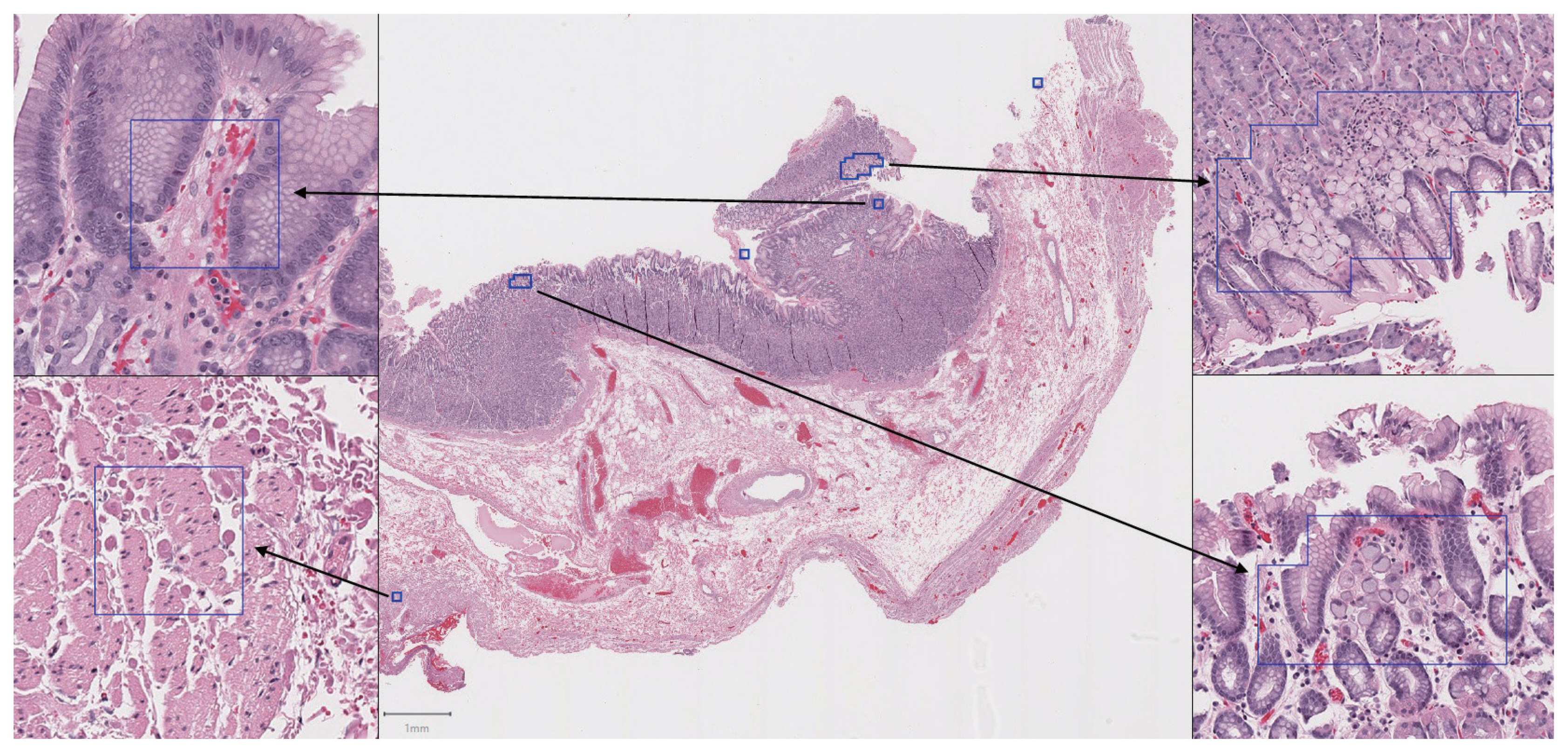
- Our second major objective was to determine whether the trained model could be used to effectively analyze whole slide images with an acceptable sensitivity and false-positive rate. On 13 whole slide images from the test cases, the sensitivity for tumor patches was 100% (24 out of 24 tumor foci overlapped with at least one patch predicted positive by the model). An example of the model’s output following analysis of a whole slide image is illustrated in Fig. 5.
- On average, false-positive results accounted for 0.098% of the non-cancer slide area (ranging from 0% to 0.17%). This was equivalent to a mean of 0.53 mm2 (approximately 0.14 100 × microscope fields) of false-positive area per slide (ranging from 0 to 0.91 mm2). In other words, the model correctly eliminated more than 99.9% of the non-cancer area from whole slide images, while correctly identifying all tumor foci present in the testing data. However, 12 out of 13 whole slide images from the test set had at least one false-positive region.
RESULTS
- In this study, we created the first dataset of annotated digital histopathology images from patients with hereditary diffuse gastric cancer. We used this data to train a CNN with DenseNet-169 architecture to accurately classify individual patches of cancer versus patches of normal background gastric tissue. The model’s performance on this task was consistent when it was evaluated on a small external validation dataset. Additionally, we applied the trained model to the far more difficult problem of analyzing whole slide images from a test set of cases that were not seen during training. On whole slide images, the model identified all tumor foci with a relatively low false-positive rate.
- The trained model performed exceptionally well on the classification of individual image patches (including images from our institution and external validation images). Its success on this task suggests that the classification of these patches is usually relatively easy. Any pathologist looking at the images in Fig. 4 would have little trouble determining which contained cancer or normal tissue, and the trained model likewise seems to have little difficulty with this distinction. For a human pathologist, the task becomes much more difficult when foci of cancer are presented in the context of a series of whole slide images from a total gastrectomy. This requires hours of sustained pathologist attention while scanning every part of the sampled tissue on each of hundreds of slides, typically at 100 × magnification. Unlike a human, a computer can scan every cell of every slide at 200× magnification with no fatigue or loss of concentration. In our analysis, this resulted in 100% sensitivity for tumor foci.
- However, as illustrated in Fig. 5, the trained model has its own set of difficulties when it analyzes a whole slide image. The number of images of normal background tissue in the training dataset is relatively small compared to the number of images of normal tissue encountered when scanning an entire whole slide image. Because a machine learning algorithm can only make predictions based on images that it has encountered during training, it will inevitably encounter areas in a whole slide image that are unlike images it has seen before, and therefore more difficult to classify. Practically, this results in occasional false-positive patches, which were present on almost every slide analyzed. The false-positive results accounted for a small portion of the total slide area, equivalent to an average of 0.14 100 × microscope fields per slide, but these false-positive regions would require interpretation by a pathologist following automated computer analysis.
- A major barrier to the implementation of an automated slide analysis system is the digitization of all relevant slides. This is a time-consuming and expensive process, but some centers have begun to routinely digitize all surgical cases, encouraging pathologists to routinely sign out cases using digital images [13]. Even after digitizing slides, automated analysis itself can be slow, requiring between 1 and 2 hours per slide in the current study. This time could be significantly improved with more powerful hardware, but remains an important consideration when assessing the potential benefits of computer-assisted diagnosis.
- Despite the successes of CNN models in pathology, there are significant barriers to their implementation. Effective training requires a large number of images containing the lesion of interest. The most successful studies analyzing whole slide images have used massive training slide datasets to maximize the experience of the model during training and minimize false-positives during testing. For example, Campanella et al. [4] utilized 44,732 whole slide images, representing a dataset many orders of magnitude greater than that used in the current study. While such an approach is undeniably impressive, it is less practical in the case of rare diseases like hereditary diffuse gastric cancer. Furthermore, most approaches require that lesional areas are manually annotated with “ground truth” labels prior to training. This is particularly time consuming in hereditary diffuse gastric cancer because of the multifocal, poorly circumscribed nature of the lesions. To our knowledge, our dataset currently represents the only fully annotated set of digital images of hereditary diffuse gastric cancer, meaning that approaches requiring larger datasets are not yet feasible.
- While the external validation data from the current study suggests that the model may generalize well to image at other institutions, it must be acknowledged that the external validation dataset we used was quite small. This reflects the difficulty of creating usable data for cases of hereditary diffuse gastric cancer, as no large-scale datasets currently exist. The model would undoubtedly benefit from being trained on a larger volume of data generated from several different labs. The current work clearly demonstrates the potential utility of deep learning in the context of hereditary diffuse gastric cancer, and further refinements as additional datasets become available in the future will continue to improve on the baseline we have established here.
- It should also be noted that the use of DenseNet-169 architecture in the current study is somewhat arbitrary. While this architecture is well known and compares favorably to other common network architectures in digital pathology [9,10], there is no reason to think that a model using another popular architecture would not be similarly successful. Because accuracy on the current task was so high, a comparison between network architectures would likely not be informative. This is not to suggest that DenseNet is necessarily superior, but only to show that deep learning in general is well suited to addressing the issue of hereditary diffuse gastric cancer.
- This model, trained on a relatively small set of images, shows encouraging progress towards computer-assisted diagnosis of hereditary diffuse gastric cancer. The case of hereditary diffuse gastric cancer may represent an ideal example of the value and effectiveness of computer-assisted diagnosis, as the strengths of computers (tirelessly scanning a large number of images) and pathologists (making intelligent decisions when encountering images that they have never encountered previously) complement each other. In an ideal scenario, a trained model could scan every slide from an entire gastrectomy specimen and present the pathologist with only the most suspicious areas, inevitably including some false-positives. Without scanning through hundreds of slides of each case, the pathologist could then focus their attention on high power images of only the most suspicious areas in order to determine whether cancer was present or whether additional investigations (for example, deeper levels or special stains) were required. Determining whether such a cooperative effort can in fact improve efficiency or accuracy will be the subject of future research.
DISCUSSION
Ethics Statement
Data collection and research methods were approved by the relevant research ethics boards (Nova Scotia Health Authority Research Ethics Board #1024339, Sunnybrook Health Sciences Centre Research Ethics Board #3152). Because no identifying information was collected, the requirement for informed consent was waived by the research ethics boards.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Author Contributions
Conceptualization: SAR, TA, WYH. Data curation: SAR, TA, WYH. Formal analysis: SAR. Funding acquisition: SAR, WYH. Investigation: SAR, TA, WYH. Methodology: SAR, TA, WYH. Project administration: TA, WYH. Resources: SAR, TA, WYH. Software: SAR. Supervision: TA, WYH. Validation: SAR. Visualization: SAR, TA, WYH. Writing – original draft: SAR. Writing – review & editing: SAR, TA, WYH. Approval of final manuscript: SAR, TA, WYH. Approval of final manuscript: all authors.
Funding Statement
This work was supported by funding from the Nova Scotia Health Authority Research Fund.
- 1. van der Post RS, Vogelaar IP, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet 2015; 52: 361-74. PubMedPMC
- 2. Rajpurkar P, Irvin J, Zhu K, et al. CheXnet: radiologist-level pneumonia detection on chest X-rays with deep learning Preprint at: https://arxiv.org/abs/1711.05225 . 2017.
- 3. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316: 2402-10. ArticlePubMed
- 4. Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019; 25: 1301-9. ArticlePubMedPMCPDF
- 5. Liu Y, Gadepalli K, Norouzi M, et al. Detecting cancer metastases on gigapixel pathology images Preprint at: https://arxiv.org/abs/1703.02442 . 2017.
- 6. Li C, Wang X, Liu W, Latecki LJ. DeepMitosis: mitosis detection via deep detection, verification and segmentation networks. Med Image Anal 2018; 45: 121-33. ArticlePubMed
- 7. Steiner DF, MacDonald R, Liu Y, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol 2018; 42: 1636-46. ArticlePubMedPMC
- 8. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2017; 2017: 4700-8. Article
- 9. Bianco S, Cadene R, Celona L, Napoletano P. Benchmark analysis of representative deep neural network architectures. IEEE Access 2018; 6: 64270-7. Article
- 10. Mormont R, Geurts P, Maree R. Comparison of deep transfer learning strategies for digital pathology. Proc Comput Soc IEEE Conf Comput Vis Pattern Recognit 2018; 2018: 2375-84. Article
- 11. Bankhead P, Loughrey MB, Fernandez JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017; 7: 16878.ArticlePubMedPMCPDF
- 12. Abadi M, Agarwal A, Barham P, et al. Tensorflow: large-scale machine learning on heterogeneous distributed systems [Internet] Mountain View: TensorFlow, 2015 [cited 2020 Jun 9]. Available from: http://tensorflow.org .
- 13. Hanna MG, Reuter VE, Samboy J, et al. Implementation of digital pathology offers clinical and operational increase in efficiency and cost savings. Arch Pathol Lab Med 2019; 143: 1545-55. ArticlePubMedPMCPDF
REFERENCES
Figure & Data
References
Citations

- Now and future of artificial intelligence-based signet ring cell diagnosis: A survey
Zhu Meng, Junhao Dong, Limei Guo, Fei Su, Jiaxuan Liu, Guangxi Wang, Zhicheng Zhao
Expert Systems with Applications.2026; 296: 129188. CrossRef - Development and application of deep learning-based diagnostics for pathologic diagnosis of gastric endoscopic submucosal dissection specimens
Soomin Ahn, Yiyu Hong, Sujin Park, Yunjoo Cho, Inwoo Hwang, Ji Min Na, Hyuk Lee, Byung-Hoon Min, Jun Haeng Lee, Jae J. Kim, Kyoung-Mee Kim
Gastric Cancer.2025; 28(4): 609. CrossRef - A Comprehensive Literature Review of the CDH1 Mutation and Its Role in Gastric Cancer
Malik Samardali, Jehad Samardaly, Ibrahim Shanti
Cureus.2025;[Epub] CrossRef - Deep learning for multiclass tumor cell detection in histopathology slides of hereditary diffuse gastric cancer
Robin Lomans, Valentina Angerilli, Joey Spronck, Liudmila L. Kodach, Irene Gullo, Fátima Carneiro, Rachel S. van der Post, Francesco Ciompi
iScience.2025; 28(8): 113064. CrossRef - Non-endoscopic Applications of Machine Learning in Gastric Cancer: A Systematic Review
Marianne Linley L. Sy-Janairo, Jose Isagani B. Janairo
Journal of Gastrointestinal Cancer.2024; 55(1): 47. CrossRef - Artificial intelligence applicated in gastric cancer: A bibliometric and visual analysis via CiteSpace
Guoyang Zhang, Jingjing Song, Zongfeng Feng, Wentao Zhao, Pan Huang, Li Liu, Yang Zhang, Xufeng Su, Yukang Wu, Yi Cao, Zhengrong Li, Zhigang Jie
Frontiers in Oncology.2023;[Epub] CrossRef - Deep learning of endoscopic features for the assessment of neoadjuvant therapy response in locally advanced rectal cancer
Anqi Wang, Jieli Zhou, Gang Wang, Beibei Zhang, Hongyi Xin, Haiyang Zhou
Asian Journal of Surgery.2023; 46(9): 3568. CrossRef - Preparing Data for Artificial Intelligence in Pathology with Clinical-Grade Performance
Yuanqing Yang, Kai Sun, Yanhua Gao, Kuansong Wang, Gang Yu
Diagnostics.2023; 13(19): 3115. CrossRef - Using Deep Learning to Predict Final HER2 Status in Invasive Breast Cancers That are Equivocal (2+) by Immunohistochemistry
Sean A. Rasmussen, Valerie J. Taylor, Alexi P. Surette, Penny J. Barnes, Gillian C. Bethune
Applied Immunohistochemistry & Molecular Morphology.2022; 30(10): 668. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure

 E-submission
E-submission