Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 51(1); 2017 > Article
-
Original Article
KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay - Sung Hak Lee*, Arthur Minwoo Chung*, Ahwon Lee, Woo Jin Oh, Yeong Jin Choi, Youn-Soo Lee, Eun Sun Jung
-
Journal of Pathology and Translational Medicine 2017;51(1):24-31.
DOI: https://doi.org/10.4132/jptm.2016.10.03
Published online: December 25, 2016
Departments of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- Corresponding Author: Eun Sun Jung, MD Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, 222 Banpo-daero, Seocho-gu, Seoul 06591, Korea Tel: +82-2-2258-1619 Fax: +82-2-2258-1628 E-mail: esjung@catholic.ac.kr
- *Sung Hak Lee and Arthur Minwoo Chung contributed equally to this work.
© 2017 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Mutations in the KRAS gene have been identified in approximately 50% of colorectal cancers (CRCs). KRAS mutations are well established biomarkers in anti–epidermal growth factor receptor therapy. Therefore, assessment of KRAS mutations is needed in CRC patients to ensure appropriate treatment.
-
Methods
- We compared the analytical performance of the cobas test to Sanger sequencing in 264 CRC cases. In addition, discordant specimens were evaluated by 454 pyrosequencing.
-
Results
- KRAS mutations for codons 12/13 were detected in 43.2% of cases (114/264) by Sanger sequencing. Of 257 evaluable specimens for comparison, KRAS mutations were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% positive percent agreement (PPA) and 91.0% negative percent agreement (NPA) for codons 12/13. Results from the cobas test and Sanger sequencing were discordant for 20 cases (7.8%). Twenty discrepant cases were subsequently subjected to 454 pyrosequencing. After comprehensive analysis of the results from combined Sanger sequencing–454 pyrosequencing and the cobas test, PPA was 97.5% and NPA was 100%.
-
Conclusions
- The cobas test is an accurate and sensitive test for detecting KRAS-activating mutations and has analytical power equivalent to Sanger sequencing. Prescreening using the cobas test with subsequent application of Sanger sequencing is the best strategy for routine detection of KRAS mutations in CRC.
- Selection of patients and tumor samples
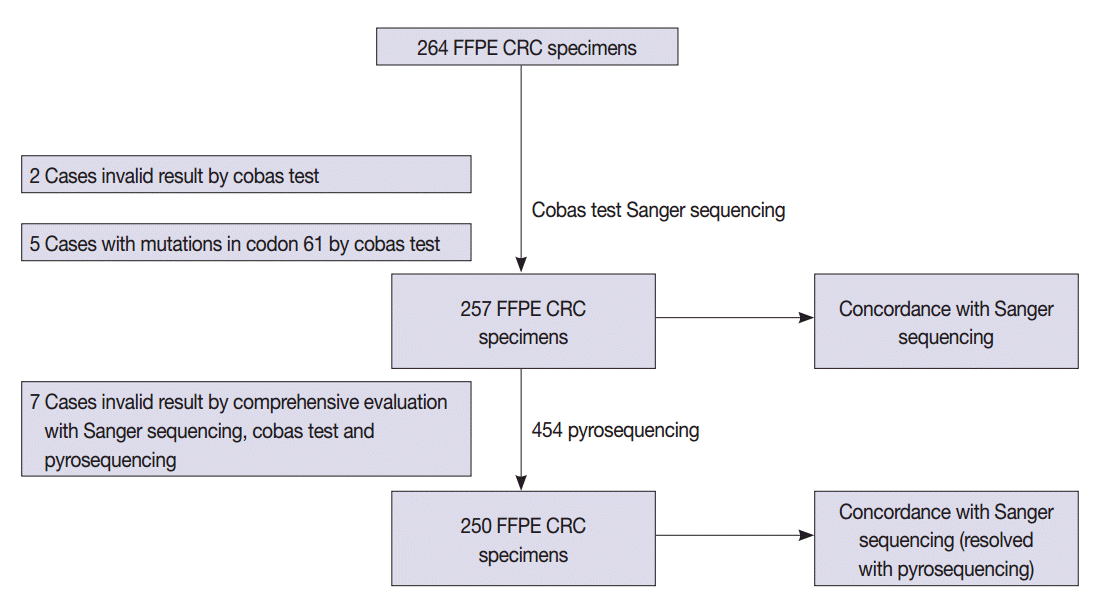
- A total of 264 patients with CRC who had undergone radical surgery at Seoul St. Mary’s Hospital, The Catholic University of Korea between 2008 and 2010 were enrolled in this study. All cases were sporadic without any family history of CRC and were examined by a pathologist who specializes in gastrointestinal tract pathology. The formalin-fixed, paraffin-embedded (FFPE) tissue samples from CRC patients were tested in accordance with protocols approved by the Institutional Review Board of The Catholic University of Korea (KC12SISI0705). Estimated tumor content ranged from 50% to 90%. The study scheme is summarized in Fig. 1.
- Direct sequencing technique for KRAS mutation
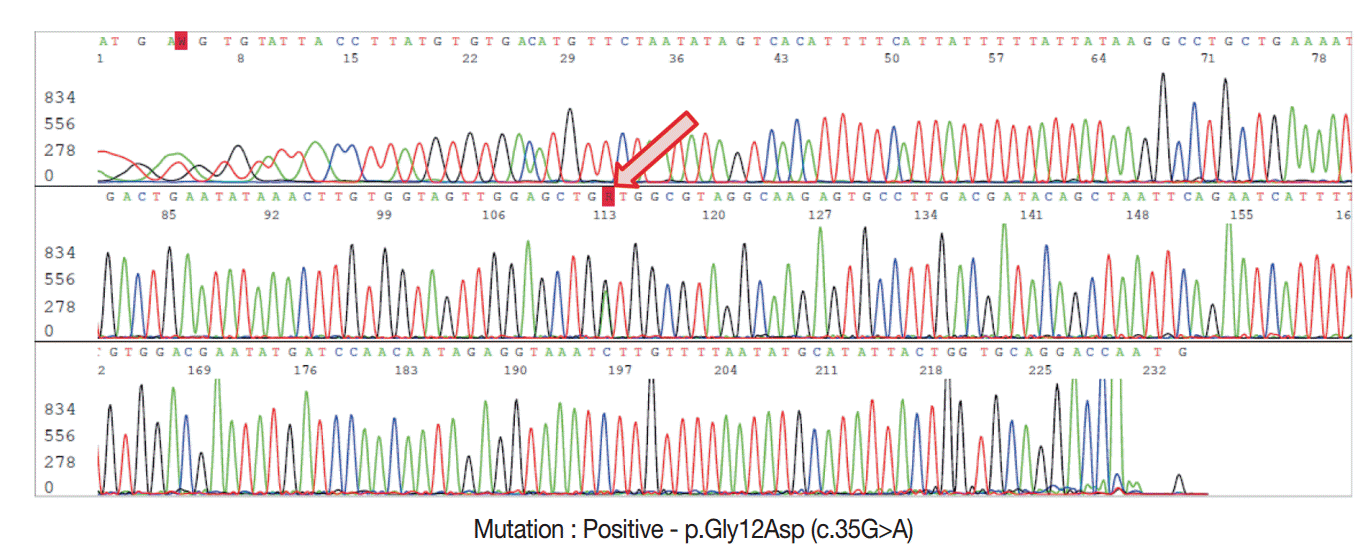
- For DNA isolation, 10-μm-thick sections from FFPE tissue samples were used for each case. The hematoxylin and eosin sections used as references were marked with a pen to indicate the tumor-rich area, and the tumor area was scraped off with a scalpel under a dissecting microscope. For genomic DNA extraction, we used the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to manufacturer’s recommendations. DNA yields were quantified using a Nanodrop spectrophotometer ND-1000 (Thermo Fisher Scientific Inc., Waltham, MA, USA). Sanger sequencing was performed using an ABI 3730 automated sequencer (Applied Biosystems Inc., Foster City, CA, USA) to detect the presence of KRAS exon 2 mutations with previously reported primers [7]. The resulting PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and the appropriate protocol on the QIAcube robotic workstation. Each chromatogram was visually inspected for abnormalities (Fig. 2).
- The cobas KRAS mutation test
- The TaqMelt PCR assay cobas KRAS Mutation Test (Roche Diagnostics) was used according to manufacturer’s protocol. First, manual specimen preparation was conducted to obtain genomic DNA from FFPE CRC tissue samples (50 ng of DNA), and PCR amplification and detection of target DNA were performed on the cobas 4800 system. Data were automatically processed by the COBAS software.
- 454 Quantitative massively parallel pyrosequencing
- Specimens showing discordant cobas test and Sanger sequencing results were retested using a quantitative massively parallel pyrosequencing method (454 GS Titanium, 454 Life Sciences, Branford, CT, USA) by Roche R&D Center China (RRDCC, Shanghai, China) using a validated protocol.
- Statistical analysis
- Data were analyzed using the SPSS statistical software ver. 21.0 (IBM Corp., Armonk, NY, USA) for Windows. The analytical performance of the cobas test compared with Sanger sequencing for the detection of KRAS mutations was evaluated by positive percent agreement (PPA), negative percent agreement (NPA), positive predictive value, and negative predictive value with two-sided 95% confidence intervals.
MATERIALS AND METHODS
- Patients and samples characteristics
- The characteristics of the patients and samples are summarized in Table 1. Patients included 161 men (61.0%) and 103 women (39.0%) with a median age of 62 years (range, 32 to 93 years). Most tissue samples were from primary tumors, while the remaining 1.5% of samples (4 of 264) were metastatic. A total of 262 patients were diagnosed with adenocarcinoma and two patients were diagnosed with neuroendocrine tumor, grade 1 (carcinoid tumor) and gastrointestinal stromal tumor. The median DNA concentration was 199.59 (range, 8.35 to 1,180.87).
- Frequency of mutations in KRAS exon 2 by Sanger sequencing and the cobas test
- The detailed mutation status of 264 cases using Sanger sequencing and the cobas test is summarized in Table 2 and Supplementary Table S1. KRAS mutations in codons 12 and 13 were detected in 114 cases (43.2%) by Sanger sequencing. KRAS mutations in codons 12, 13, and 61 were revealed in 123 cases (46.6%) by the cobas test. Among the 123 mutation cases on the cobas test, five cases were revealed to have mutations in codon 61 that were not detected by Sanger sequencing, but instead only by the cobas test. Therefore, we excluded five cases of codon 61 mutation from comparison. Two cases had invalid results on the cobas test due to inadequate specimen and were excluded from comparison. In total, we evaluated 257 cases for comparison.
- Overall concordance for detecting KRAS mutation of codons 12/13 by Sanger sequencing and the cobas test
- KRAS mutations for 257 evaluable cases were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% PPA and 91.0% NPA for codons 12/13. Discordant ratio of Sanger sequencing and cobas test was 7.7% (20 out of 257 cases) (Table 3).
- Analysis for 20 discrepant cases with 454 pyrosequencing
- For further analysis of 20 discordant cases, we performed 454 pyrosequencing. Among 20 cases, seven cases showed inconclusive results due to low allele frequency on 454 pyrosequencing (Nos. 32 and 82), different mutation types among test modalities (Nos. 26, 100, and 104), and inadequate specimen for 454 pyrosequencing (Nos. 34 and 116).
- In one case (No. 54), the result of Sanger sequencing coincides with that of 454 pyrosequencing. In the remaining cases, 10 cases (Nos. 49, 80, 85, 86, 88, 103, 125, 149, 159, and 221) had no mutation with Sanger sequencing, whereas a single nucleotide variant (SNV) was identified by the cobas test and 454 pyrosequencing. In those cases, mutations with high allele frequency by 454 sequencing were regarded as the final results. In two cases (Nos. 97 and 223), no mutation was identified by the cobas test while different mutation types were shown between Sanger sequencing and 454 sequencing. Considering repetitive failure for PCR amplification or inconsistent results on the cobas test, this discrepancy might be due to extensive DNA fragmentation and/or FFPE-related artifacts. Thus, mutations with a high allele frequency by 454 sequencing were considered as the final results in these cases (Table 4, Supplementary Tables S2, S3).
- Overall concordance for detecting KRAS mutation of codons 12/13 (the cobas test and combined Sanger and 454 sequencing resolution)
- After discordant resolution with 454 pyrosequencing, we analyzed 250 cases for concordance between the cobas test and combined Sanger sequencing–454 pyrosequencing. KRAS mutations for 250 evaluable cases were detected in 118 cases (46.0%) by combined Sanger sequencing–454 pyrosequencing and 115 cases (46.0%) by the cobas test. Concordance between the cobas test and combined Sanger sequencing–454 pyrosequencing was 97.5% PPA and 100% NPA. The discordant ratio of the Sanger sequencing and the cobas test was 1.2% (3 out of 250 cases) (Table 5).
RESULTS
- There are various testing assays available for KRAS mutations, and several comparative studies on the analytical power of these methods in the clinical context have been conducted [3,8-10]. In this study, we compared the performance of two platforms for detecting KRAS-activating mutations in Korean CRC cohorts. Determination of the KRAS mutation status is now obligatory for treatment with anti-EGFR monoclonal antibodies such as cetuximab and panitumumab in patients with metastatic CRC, as only patients with wild-type KRAS may respond to treatment [11,12]. Furthermore, in patients with metastatic CRC and KRAS-activating mutations, treatment with anti-EGFR monoclonal antibodies may result in deleterious effects on progression-free survival, which highlights the necessity of maximizing the detection of KRAS mutations [13,14]. However, validated and standardized procedures for KRAS somatic mutation testing were lacking until recently. Sanger sequencing remains the gold standard assay for detecting all possible mutations, but is generally considered to be less sensitive than other methods [15].
- The cobas (KRAS mutation) test is a CE-IVD validated method based on real-time PCR and TaqMelt technology [16]. The accuracy of this method is similar to that of massively parallel pyrosequencing. In this study, the cobas test showed a ≥95% correct mutation call rate for the recommended DNA input of ≥50 ng. It has been designed to detect 19 common KRAS mutations in codons 12, 13, and 61 in FFPE-derived DNA from CRC samples [17]. Moreover, the cobas test has been reported to detect mutations in tissue samples containing a minimal amount of 5% of tumor DNA and showed comparable and greater sensitivity for low-level mutant allele burden than Sanger sequencing [17]. The results indicate the presence or absence of KRAS mutations without detailed information regarding the exact mutated site. While 454 pyrosequencing also shows a sensitivity of 5%, this method did not confirm the results obtained from other test assays. We used 454 pyrosequencing as a reference method to evaluate discrepant results between the cobas test and Sanger sequencing.
- In this study, there were 20 discordant results (7.8%) between the cobas test and the Sanger sequencing, excluding two cases with invalid results and five cases with KRAS mutations in codon 61 by the cobas test. For further evaluation of samples with discrepant mutation results, each sample was run three times on Sanger sequencing and the cobas test. If invalid results are obtained, a new sample from a different tissue block was used if possible, which might result in different mutation types from intratumoral heterogeneity. In those cases, we made a great effort to choose the tissue section with the higher tumor portions (>70%), and no necrotic or mucinous areas.
- In five cases (Nos. 26, 97, 100, 104, and 223), different mutation types were identified between Sanger sequencing and 454 sequencing. In addition, two cases (Nos. 32 and 82) showed low allele frequency on 454 pyrosequencing. Considering repetitive failure for PCR amplification in Sanger sequencing and/or invalid results in the cobas test, this discrepancy among test modalities might come from extensive DNA fragmentation from old formalin-fixed tissues. FFPE-related artifacts, i.e., spontaneous deamination of cytosine bases, might have a great effect on this inconsistency. Recently, uracil lesions in which hydrolytic deamination of cytosine bases to uracil takes place have been identified as major sources of sequence artifacts in FFPE DNA, leading to artifactual C:G>T:A variants [18]. These sequence artifacts are known to be detected more frequently when low copy numbers of FFPE DNA are provided [19].
- In ten cases (Nos. 49, 80, 85, 86, 88, 103, 125, 149, 159, and 221), no mutation was detected by Sanger sequencing, whereas SNVs were identified by the cobas test and 454 sequencing. These discrepancies might result from low analytical sensitivity of Sanger sequencing. Although Sanger sequencing is considered to be the gold standard in mutation testing, it has a modest limit of detection that can be highly variable depending on the specific mutation sequence. A recent study has shown that Sanger sequencing yielded 11.1% false-positives and 6.1% false-negatives for KRAS mutation detection using an automated interpretation algorithm with a 10% threshold, highlighting the need for manual review of all Sanger sequencing data (Table 4, Supplementary Tables S2, S3) [20].
- In addition to the above mentioned factors, artifactual mutations from FFPE tissues can be caused by various sources, including over-fixation in 10% non-buffered formalin, oxidative DNA damage during sample preparation, DNA polymerase error, pseudogene amplification, sequencing chemistry, and errors of alignment and/or annotation [19,21-24]. Several studies have shown that the numbers of SNV identified in formalin-fixed tissues were higher than those in matched frozen tissues, which suggests the possibilities of artifactual sequence changes, thus increasing the risk of false-positive mutation calls [25,26]. Additionally, the discrepant results may have also come from uncommon KRAS mutations that cannot be detected using the cobas design. Understanding these issues is important for accurate interpretation of various mutations detected.
- Although a “mutated in codon 12/13 or 61” result in the cobas KRAS mutation test is a sufficient basis for the application of anti-EGFR monoclonal antibodies in mCRC, several studies suggest that different mutation sites have different clinical impacts on cetuximab and/or panitumumab efficacy. The potential differential sensitivity of KRAS G13D mutation to anti-EGFR monoclonal antibodies has been reported in a small number of studies, suggesting that the addition of cetuximab may be beneficial to CRC patients with a KRAS G13D mutation [27,28]. However, no significant difference between KRAS G13D and other KRAS mutated CRCs was detected in terms of treatment benefit from anti-EGFR monoclonal antibodies for mCRC [29].
- Detection of KRAS-activating mutations in CRC has largely only focused on mutations in codons 12 and 13. However, recent data has shown that clinicopathological features and gene expression profiles of CRCs harboring non-traditional KRAS mutations appear to be similar to those of tumors with KRAS mutations in codons 12 and 13 [30]. KRAS mutations in codon 61 and/or 146 are additional hotspots in CRC, and the available data from a small number of studies suggest that resistance to anti-EGFR therapy is associated with KRAS mutation at these sites [31,32]. Pre-screening using the cobas KRAS mutation test, which accurately detects all main KRAS mutations in codons 12, 13, and 61, with further evaluation by Sanger sequencing enables the identification of codon 12 and 13 mutations. This may be an easy and reliable approach for routine diagnostic purposes regarding KRAS mutations in mCRC. In addition, combining both mutation assays greatly reduces the probability of obtaining false-negative or false-positive results.
- This study also emphasized that the preanalytical step must be strictly controlled because DNA degradation was the main cause of “not available” results, which leads to non-conclusive results, and samples cannot be evaluated using other genetic tests.
- Several techniques for detecting KRAS mutations are currently available, but there is limited data supporting the analytical performance of individual methods compared with other methods [33-35]. In addition, recent studies demonstrated the variability of mutation tests resulting from different clinical laboratories [36]. In contrast to other KRAS mutation detecting kits, such as the Therascreen KRAS mutation test, the cobas test can detect 12 additional mutation sites, representing approximately 1% of all KRAS mutation cases of CRC based on data from the Catalogue of Somatic Mutations in Cancer (COSMIC) database [3]. However, several studies revealed that KRAS mutations in codon 61 are more prevalent than reflected by the COSMIC data in CRC [3,37,38]. Thus, more comprehensive KRAS mutation coverage may be helpful for selecting patients for anti-EGFR target therapies.
- A limitation of this study is that the study design was retrospective and the sample size was unsatisfactory to evaluate the significance of infrequent KRAS mutation subtypes. Subsequent translational prospective studies from different cohorts are needed to confirm our data. In addition, some invalid and discrepant results were not sufficiently resolved. Nevertheless, we demonstrated that the cobas KRAS mutation test is a reproducible companion diagnostic test in patients with CRCs under consideration for anti-EGFR target therapies when used with Sanger sequencing.
- In conclusion, we confirmed that the cobas KRAS mutation test is an accurate and sensitive test for detecting KRAS-activating mutations and has equivalent analytical power to the Sanger direct sequencing method. Therefore, pre-screening using the cobas KRAS mutation assay with subsequent application of Sanger sequencing is the best strategy for routine diagnostic purposes regarding KRAS mutations in mCRC.
DISCUSSION
Electronic Supplementary Material
| Sanger sequencing | The cobas test | |
|---|---|---|
| Mutation | ||
| Codon 12 | 90 (34.1) | 118 (44.7) |
| Codon 13 | 24 (9.1) | |
| Codon 61 | - | 5 (1.9) |
| No mutation | 150 (56.8) | 139 (52.7) |
| N/A specimen | 0 | 2 (0.8) |
| Total | 264 (100) | 264 (100) |
| The cobas test |
Sanger sequencing |
||
|---|---|---|---|
| Mutation detected | Mutation not detected | Total | |
| Mutation detected | 105 (40.9) | 13 (5.1) | 118 (45.9) |
| Mutation not detected | 7 (2.7) | 132 (51.3) | 139 (54.1) |
| Total | 112 (43.6) | 145 (56.4) | 257a (100) |
Values are presented as number (%). Positive percent agreement (sensitivity): 93.8% (95% confidence interval [CI], 87.6 to 97.5). Negative percent agreement (specificity): 91.0% (95% CI, 85.2 to 95.1). Positive predictive value: 89.0% (95% CI, 81.9 to 94.0). Negative predictive value: 95.0% (95% CI, 89.9 to 98.0). Positive likelihood ratio: 10.5 (95% CI, 6.2 to 17.6). Negative likelihood ratio: 0.1 (95% CI, 0.0 to 0.1).
aOf 264 cases, two cases showed invalid result and five cases revealed KRAS mutations in codon 61 by the cobas test.
| The Cobas test |
Sanger sequencing and 454 pyrosequencing |
||
|---|---|---|---|
| Mutation detected | Mutation not detected | Total | |
| Mutation detected | 115 (46.0) | 0 | 115 (46.0) |
| Mutation not detected | 3 (1.2) | 132 (52.8) | 135 (54.0) |
| Total | 118 (47.2) | 132 (52.8) | 250a (100) |
Values are presented as number (%). Positive percent agreement (sensitivity): 97.5% (95% confidence interval [CI], 92.8 to 99.5). Negative percent agreement (specificity): 100% (95% CI, 97.2 to 100). Positive predictive value: 100% (95% CI, 96.9 to 100). Negative predictive value: 97.8% (95% CI, 93.7 to 99.5). Negative likelihood ratio: 0.03 (95% CI, 0.0 to 0.1).
aOf 257 cases, seven cases showed invalid result by comprehensive evaluation with Sanger sequencing, the cobas test and 454 pyrosequencing.
- 1. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer 2003; 3: 459-65. ArticlePubMedPDF
- 2. Bos JL. Ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682-9. PubMed
- 3. Lee S, Brophy VH, Cao J, et al. Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch 2012; 460: 141-9. ArticlePubMed
- 4. Venook AP. Epidermal growth factor receptor-targeted treatment for advanced colorectal carcinoma. Cancer 2005; 103: 2435-46. ArticlePubMed
- 5. Dahabreh IJ, Terasawa T, Castaldi PJ, Trikalinos TA. Systematic review: anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med 2011; 154: 37-49. ArticlePubMed
- 6. Querings S, Altmüller J, Ansén S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One 2011; 6: e19601.ArticlePubMedPMC
- 7. Kim SY, Choi EJ, Yun JA, et al. Syndecan-1 expression is associated with tumor size and EGFR expression in colorectal carcinoma: a clinicopathological study of 230 cases. Int J Med Sci 2015; 12: 92-9. ArticlePubMedPMC
- 8. Harlé A, Busser B, Rouyer M, et al. Comparison of COBAS 4800 KRAS, TaqMan PCR and high resolution melting PCR assays for the detection of KRAS somatic mutations in formalin-fixed paraffin embedded colorectal carcinomas. Virchows Arch 2013; 462: 329-35. ArticlePubMedPDF
- 9. Sharma A, Zhang G, Aslam S, Yu K, Chee M, Palma JF. Novel approach for clinical validation of the cobas KRAS mutation test in advanced colorectal cancer. Mol Diagn Ther 2016; 20: 231-40. ArticlePubMedPMCPDF
- 10. Gonzalez de Castro D, Angulo B, Gomez B, et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer 2012; 107: 345-51. ArticlePubMedPMCPDF
- 11. Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26: 1626-34. ArticlePubMed
- 12. Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009; 360: 1408-17. ArticlePubMed
- 13. Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the firstline treatment of metastatic colorectal cancer. J Clin Oncol 2009; 27: 663-71. ArticlePubMed
- 14. Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010; 28: 4697-705. ArticlePubMed
- 15. Malapelle U, Carlomagno C, Salatiello M, et al. KRAS mutation detection by high-resolution melting analysis significantly predicts clinical benefit of cetuximab in metastatic colorectal cancer. Br J Cancer 2012; 107: 626-31. ArticlePubMedPMCPDF
- 16. Malapelle U, Carlomagno C, de Luca C, Bellevicine C, Troncone G. KRAS testing in metastatic colorectal carcinoma: challenges, controversies, breakthroughs and beyond. J Clin Pathol 2014; 67: 1-9. ArticlePubMed
- 17. Angulo B, Lopez-Rios F, Gonzalez D. A new generation of companion diagnostics: cobas BRAF, KRAS and EGFR mutation detection tests. Expert Rev Mol Diagn 2014; 14: 517-24. ArticlePubMed
- 18. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 2015; 61: 64-71. ArticlePubMedPDF
- 19. Sah S, Chen L, Houghton J, et al. Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome Med 2013; 5: 77.ArticlePubMedPMCPDF
- 20. Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 2010; 12: 425-32. PubMedPMC
- 21. Costello M, Pugh TJ, Fennell TJ, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 2013; 41: e67.ArticlePubMedPMC
- 22. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011; 108: 9530-5. ArticlePubMedPMC
- 23. Xuan J, Yu Y, Qing T, Guo L, Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett 2013; 340: 284-95. ArticlePubMed
- 24. Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 2008; 18: 1851-8. ArticlePubMedPMC
- 25. Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol 2004; 4: 1.ArticlePubMedPMCPDF
- 26. Wong SQ, Li J, Tan AY, et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics 2014; 7: 23.ArticlePubMedPMCPDF
- 27. De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 2010; 304: 1812-20. ArticlePubMed
- 28. Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res 2007; 67: 2643-8. ArticlePubMedPDF
- 29. Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012; 486: 532-6. ArticlePubMedPMCPDF
- 30. Morris VK, Lucas FA, Overman MJ, et al. Clinicopathologic characteristics and gene expression analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal cancer. Ann Oncol 2014; 25: 2008-14. ArticlePubMedPMC
- 31. Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 2009; 101: 715-21. ArticlePubMedPMCPDF
- 32. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010; 11: 753-62. ArticlePubMed
- 33. Gao J, Li YY, Sun PN, Shen L. Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol 2010; 16: 4858-64. ArticlePubMedPMC
- 34. Angulo B, García-García E, Martínez R, et al. A commercial realtime PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations: a morphology-based approach in colorectal carcinoma. J Mol Diagn 2010; 12: 292-9. PubMedPMC
- 35. Whitehall V, Tran K, Umapathy A, et al. A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn 2009; 11: 543-52. ArticlePubMedPMC
- 36. Beau-Faller M, Degeorges A, Rolland E, et al. Cross-validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non-small cell lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR-TKIs in non-small cell lung carcinoma [ERMETIC] project- -part 1). J Thorac Oncol 2011; 6: 1006-15. ArticlePubMed
- 37. Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 2009; 27: 5931-7. ArticlePubMed
- 38. Gaedcke J, Grade M, Jung K, et al. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol 2010; 94: 76-81. ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
Journal of Pathology and Translational Medicine.2023; 57(4): 217. CrossRef - Assessment of KRAS and NRAS status in metastatic colorectal cancer: Experience of the National Institute of Oncology in Rabat Morocco
Chaimaa Mounjid, Hajar El Agouri, Youssef Mahdi, Abdelilah Laraqui, En-nacer Chtati, Soumaya Ech-charif, Mouna Khmou, Youssef Bakri, Amine Souadka, Basma El Khannoussi
Annals of Cancer Research and Therapy.2022; 30(2): 80. CrossRef - The current understanding on the impact of KRAS on colorectal cancer
Mingjing Meng, Keying Zhong, Ting Jiang, Zhongqiu Liu, Hiu Yee Kwan, Tao Su
Biomedicine & Pharmacotherapy.2021; 140: 111717. CrossRef - Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer
Barbora Vanova, Michal Kalman, Karin Jasek, Ivana Kasubova, Tatiana Burjanivova, Anna Farkasova, Peter Kruzliak, Dietrich Busselberg, Lukas Plank, Zora Lasabova
Clinical and Experimental Medicine.2019; 19(2): 219. CrossRef - CRISPR Technology for Breast Cancer: Diagnostics, Modeling, and Therapy
Rachel L. Mintz, Madeleine A. Gao, Kahmun Lo, Yeh‐Hsing Lao, Mingqiang Li, Kam W. Leong
Advanced Biosystems.2018;[Epub] CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
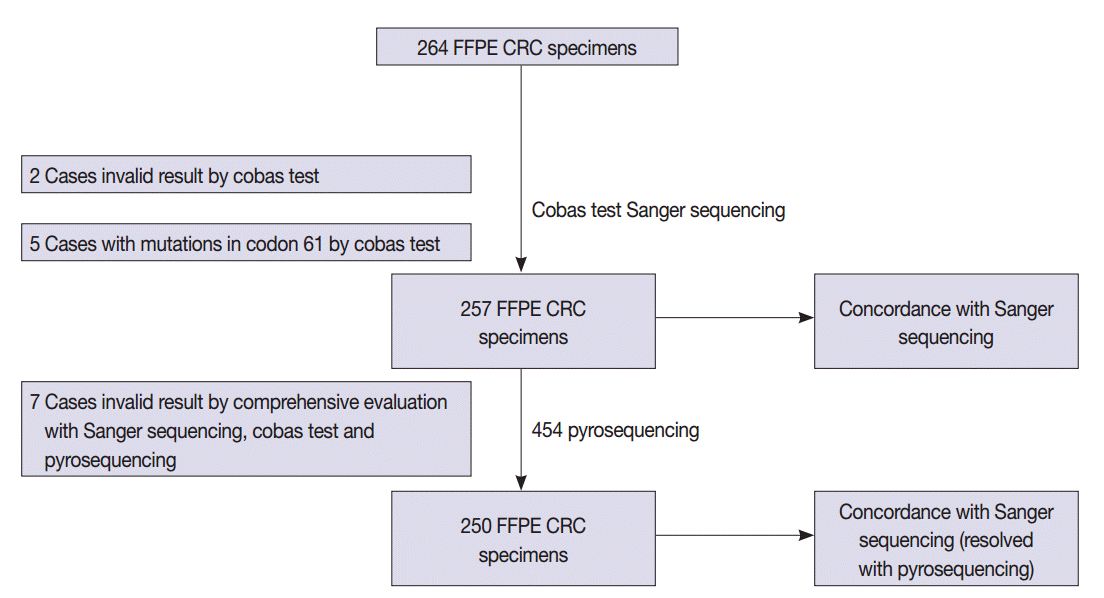
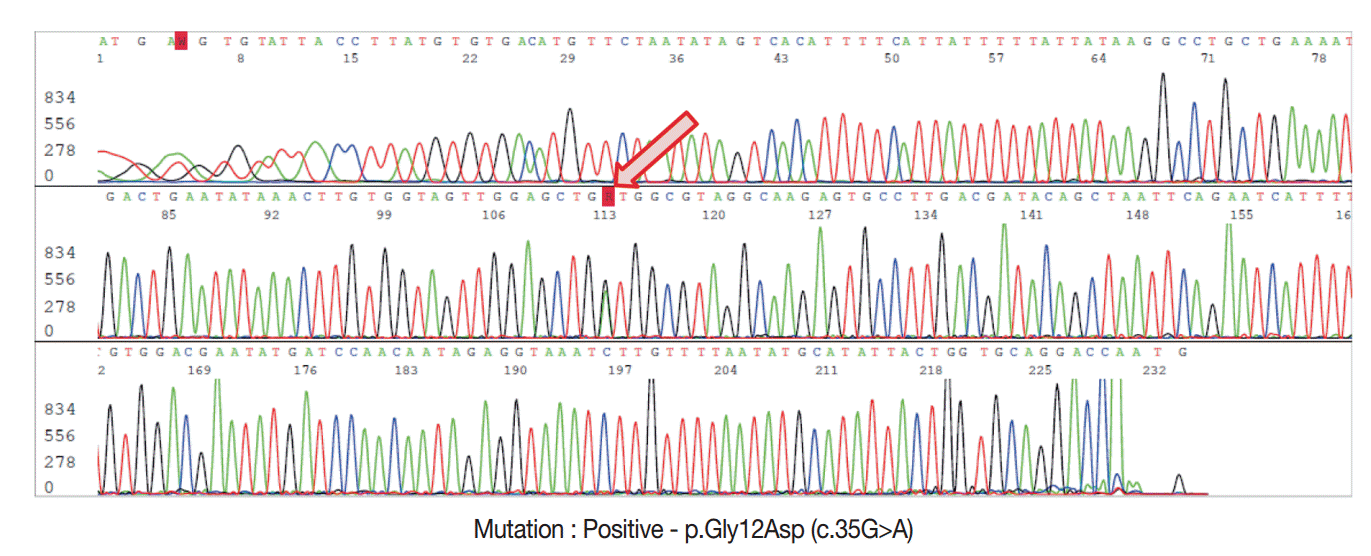
Fig. 1.
Fig. 2.
| Characteristic | No. (%) |
|---|---|
| Total patients | 264 |
| Age, median (range, yr) | 62 (32–93) |
| Gender | |
| Male | 161 (61.0) |
| Female | 103 (39.0) |
| Sites | |
| Primary | 260 (98.5) |
| Metastasis | 4 (1.5) |
| DNA concentration (ng/µL) | 199.59 (8.35–1,180.87) |
| DNA purity (260/280) | 1.99 (1.59–3.01) |
| Sanger sequencing | The cobas test | |
|---|---|---|
| Mutation | ||
| Codon 12 | 90 (34.1) | 118 (44.7) |
| Codon 13 | 24 (9.1) | |
| Codon 61 | - | 5 (1.9) |
| No mutation | 150 (56.8) | 139 (52.7) |
| N/A specimen | 0 | 2 (0.8) |
| Total | 264 (100) | 264 (100) |
| The cobas test | Sanger sequencing |
||
|---|---|---|---|
| Mutation detected | Mutation not detected | Total | |
| Mutation detected | 105 (40.9) | 13 (5.1) | 118 (45.9) |
| Mutation not detected | 7 (2.7) | 132 (51.3) | 139 (54.1) |
| Total | 112 (43.6) | 145 (56.4) | 257 |
| Sample ID | Sanger sequencing | The cobas test | 454 Pyrosequencing |
Combined result | |
|---|---|---|---|---|---|
| Variant | Frequency | ||||
| 026 | G12D | Not detected | G13D | 0.65 | N/A |
| 054 | G12D | Not detected | G12D | 0.69 | G12D |
| 082 | G12D | Not detected | G12D/G12V | 0.68/1.78 | N/A |
| 097 | G12D | Not detected | G12V | 10.92 | G12V |
| 100 | G12D | Not detected | G12A/G12S/G12V | 3.13/0.45/13.2 | N/A |
| 104 | G13D | Not detected | G12D/G12S/G12V/G13D | 1.06/0.79/3.62/0.36 | N/A |
| 223 | G12D | Not detected | G12V | 17.58 | G12V |
| 032 | Negative | Codon 12/13 | G12C/G13D | 2.43/0.38 | N/A |
| 034 | Netative | Codon 12/14 | N/A | N/A | N/A |
| 049 | Negative | Codon 12/13 | G12V/G13S | 23.56/0.34 | G12V |
| 080 | Negative | Codon 12/13 | G12C/G12V/G13D/G13S | 15.25/6.5/1.62/1.29 | G12C |
| 085 | Negative | Codon 12/13 | G12D/G12V | 4.2/13.53 | G12V |
| 086 | Negative | Codon 12/13 | G12V/G13D | 9.72/1.09 | G12V |
| 088 | Negative | Codon 12/13 | G12V | 17.5 | G12V |
| 103 | Negative | Codon 12/13 | G12D/G12V/G13D | 1.86/4.47/0.81 | G12V |
| 116 | Negative | Codon 12/13 | N/A | N/A | N/A |
| 125 | Negative | Codon 12/13 | G12V/G13D | 15.66/3.88 | G12V |
| 149 | Negative | Codon 12/13 | G12V | 24.9 | G12V |
| 159 | Negative | Codon 12/13 | G12D/G12S/G12V | 0.4/1.41/6.7 | G12V |
| 221 | Negative | Codon 12/13 | G12V/G13S | 8.92/1.64 | G12V |
| The Cobas test | Sanger sequencing and 454 pyrosequencing |
||
|---|---|---|---|
| Mutation detected | Mutation not detected | Total | |
| Mutation detected | 115 (46.0) | 0 | 115 (46.0) |
| Mutation not detected | 3 (1.2) | 132 (52.8) | 135 (54.0) |
| Total | 118 (47.2) | 132 (52.8) | 250 |
Values are presented as number (%). N/A, not available.
Values are presented as number (%). Positive percent agreement (sensitivity): 93.8% (95% confidence interval [CI], 87.6 to 97.5). Negative percent agreement (specificity): 91.0% (95% CI, 85.2 to 95.1). Positive predictive value: 89.0% (95% CI, 81.9 to 94.0). Negative predictive value: 95.0% (95% CI, 89.9 to 98.0). Positive likelihood ratio: 10.5 (95% CI, 6.2 to 17.6). Negative likelihood ratio: 0.1 (95% CI, 0.0 to 0.1). Of 264 cases, two cases showed invalid result and five cases revealed
N/A, not available.
Values are presented as number (%). Positive percent agreement (sensitivity): 97.5% (95% confidence interval [CI], 92.8 to 99.5). Negative percent agreement (specificity): 100% (95% CI, 97.2 to 100). Positive predictive value: 100% (95% CI, 96.9 to 100). Negative predictive value: 97.8% (95% CI, 93.7 to 99.5). Negative likelihood ratio: 0.03 (95% CI, 0.0 to 0.1). Of 257 cases, seven cases showed invalid result by comprehensive evaluation with Sanger sequencing, the cobas test and 454 pyrosequencing.

 E-submission
E-submission




