Search
- Page Path
- HOME > Search
- Clinicopathological and molecular mechanisms of CLDN18.2 in gastric cancer aggressiveness: a high-risk population study with multi-omics profiling
- Hengquan Wu, Mei Li, Gang Wang, Peiqing Liao, Peng Zhang, Luxi Yang, Yumin Li, Tao Liu, Wenting He
- J Pathol Transl Med. 2026;60(1):47-57. Published online January 5, 2026
- DOI: https://doi.org/10.4132/jptm.2025.09.11
- 669 View
- 81 Download
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
The tight junction protein claudin18.2 (CLDN18.2) has been implicated in poor prognosis and suboptimal immunotherapy response in gastric cancer (GC). This study investigates the clinicopathological relevance of CLDN18.2 expression and its association with molecular subtypes in GC patients from a high-incidence region, combining transcriptomic and proteomic approaches to explore how CLDN18.2 contributes to progression and metastasis.
Methods
A retrospective cohort of 494 GC patients (2019–2024) underwent immunohistochemical analysis for CLDN18.2, Epstein-Barr virus (Epstein–Barr virus–encoded RNA), p53, human epidermal growth factor receptor 2 (HER2), and mismatch repair proteins (MLH1, MSH2, PMS2, and MSH6). CLDN18.2 positivity was defined as moderate to strong (2+/3+) membranous staining in ≥75% of tumor cells. Clinicopathological correlations, biomarker associations, and survival outcomes were evaluated. Transcriptomic and proteomic sequencing was performed to explore molecular mechanisms.
Results
CLDN18.2 positivity was observed in 26.9% (133/494) of gastric adenocarcinomas. CLDN18.2-positive tumors correlated with TNM stage (p = .003) and shorter overall survival (p = .018). No associations were identified with age, sex, HER2 status, microsatellite instability, or Epstein-Barr virus infection. Transcriptomic profiling revealed CLDN18.2-high tumors enriched in pathways involving cell junction disruption, signaling regulation, and immune modulation. Proteomic profiling showed that tumors with high CLDN18.2 were enriched in multiple mechanism-related pathways such as integrated metabolic reprogramming, cytoskeletal recombination, immune microenvironment dysregulation, and pro-survival signaling. These mechanisms may collectively contribute to tumor progression and metastasis.
Conclusions
CLDN18.2 overexpression is associated with poor prognosis in GC patients. Transcriptomic and proteomic analyses demonstrate that CLDN18.2 promotes tumor progression and metastasis, underscoring its potential as an independent prognostic factor in regions with a high incidence of GC.
- Next step of molecular pathology: next-generation sequencing in cytology
- Ricella Souza da Silva, Fernando Schmitt
- J Pathol Transl Med. 2024;58(6):291-298. Published online November 7, 2024
- DOI: https://doi.org/10.4132/jptm.2024.10.22
- 6,036 View
- 370 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - The evolving landscape of precision oncology underscores the pivotal shift from morphological diagnosis to treatment decisions driven by molecular profiling. Recent guidelines from the European Society for Medical Oncology recomend the use of next-generation sequencing (NGS) across a broader range of cancers, reflecting its superior efficiency and clinical value. NGS not only updates oncology testing by offering quicker, sample-friendly, and sensitive analysis but also reduces the need for multiple individual tests. Cytology samples, often obtained through less invasive methods, can yield high-quality genetic material suitable for molecular analysis. This article focuses on optimizing the use of cytology samples in NGS, and outlines their potential benefits in identifying actionable molecular alterations for targeted therapies across various solid tumors. It also addresses the need for validation studies and the strategies to incorporate or combine different types of samples into routine clinical practice. Integrating cytological and liquid biopsies into routine clinical practice, alongside conventional tissue biopsies, offers a comprehensive approach to tumor genotyping, early disease detection, and monitoring of therapeutic responses across various solid tumor types. For comprehensive biomarker characterization, all patient specimens, although limited, is always valuable.
-
Citations
Citations to this article as recorded by- The World Health Organization Reporting System for Lymph Node, Spleen, and Thymus Cytopathology: Part 1 – Lymph Node
Immacolata Cozzolino, Mats Ehinger, Maria Calaminici, Andrea Ronchi, Mousa A. Al-Abbadi, Helena Barroca, Beata Bode-Lesniewska, David F. Chhieng, Ruth L. Katz, Oscar Lin, L. Jeffrey Medeiros, Martha Bishop Pitman, Arvind Rajwanshi, Fernando C. Schmitt, Ph
Acta Cytologica.2025; : 1. CrossRef - The impact of cytological preparation techniques on RNA quality: A comparative study on smear samples
Cisel Aydin Mericoz, Gulsum Caylak, Elif Sevin Sanioglu, Zeynep Seçil Satilmis, Ayse Humeyra Dur Karasayar, Ibrahim Kulac
Cancer Cytopathology.2025;[Epub] CrossRef - Reimagining cytopathology in the molecular era: Integration or fragmentation?
Sumanta Das, R. Naveen Kumar, Biswajit Dey, Pranjal Kalita
Cytojournal.2025; 22: 94. CrossRef
- The World Health Organization Reporting System for Lymph Node, Spleen, and Thymus Cytopathology: Part 1 – Lymph Node
- Colorectal cancer with a germline BRCA1 variant inherited paternally: a case report
- Kyoung Min Kim, Min Ro Lee, Ae Ri Ahn, Myoung Ja Chung
- J Pathol Transl Med. 2024;58(6):341-345. Published online September 5, 2024
- DOI: https://doi.org/10.4132/jptm.2024.08.14
- 6,772 View
- 306 Download
-
 Abstract
Abstract
 PDF
PDF - BRCA genes have well-known associations with breast and ovarian cancers. However, variations in the BRCA gene, especially germline variations, have also been reported in colorectal cancer (CRC). We present the case of a rectal cancer with a germline BRCA1 variation inherited from the paternal side. A 39-year-old male was admitted with rectal cancer. The patient underwent surgical resection and the pathologic diagnosis was adenocarcinoma. Next-generation sequencing was performed and a BRCA1 variant was detected. Reviewing the public database and considering the young age of the patient, the variant was suggested to be germline. The patient’s father had had prostate cancer and next-generation sequencing testing revealed an identical BRCA1 variant. In the BRCA cancer group, there is relatively little attention paid to male cancers. The accumulation of male CRC cases linked to BRCA variations may help clarify the potential pathological relationship between the two.
- Concurrent intestinal plasmablastic lymphoma and diffuse large B-cell lymphoma with a clonal relationship: a case report and literature review
- Nao Imuta, Kosuke Miyai, Motohiro Tsuchiya, Mariko Saito, Takehiro Sone, Shinichi Kobayashi, Sho Ogata, Fumihiko Kimura, Susumu Matsukuma
- J Pathol Transl Med. 2024;58(4):191-197. Published online June 25, 2024
- DOI: https://doi.org/10.4132/jptm.2024.05.14
- 4,729 View
- 223 Download
-
 Abstract
Abstract
 PDF
PDF - Herein, we report a case of plasmablastic lymphoma (PBL) and diffuse large B-cell lymphoma (DLBCL) that occurred concurrently in the large intestine. An 84-year-old female presented with a palpable rectal tumor and ileocecal tumor observed on imaging analyses. Endoscopic biopsy of both lesions revealed lymphomatous round cells. Hartmann’s operation and ileocecal resection were performed for regional control. The ileocecal lesion consisted of a proliferation of CD20/CD79a-positive lymphoid cells, indicative of DLBCL. In contrast, the rectal tumor showed proliferation of atypical cells with pleomorphic nuclei and abundant amphophilic cytoplasm, with immunohistochemical findings of CD38/CD79a/MUM1/MYC (+) and CD20/CD3/CD138/PAX5 (–). Tumor cells were positive for Epstein-Barr virus– encoded RNA based on in situ hybridization and MYC rearrangement in fluorescence in situ hybridization analysis. These findings indicated the rectal tumor was most likely a PBL. Sequencing analysis for immunoglobulin heavy variable genes indicated a common B-cell origin of the two sets of lymphoma cells. This case report and literature review provide new insights into PBL tumorigenesis.
- Clinical practice recommendations for the use of next-generation sequencing in patients with solid cancer: a joint report from KSMO and KSP
- Miso Kim, Hyo Sup Shim, Sheehyun Kim, In Hee Lee, Jihun Kim, Shinkyo Yoon, Hyung-Don Kim, Inkeun Park, Jae Ho Jeong, Changhoon Yoo, Jaekyung Cheon, In-Ho Kim, Jieun Lee, Sook Hee Hong, Sehhoon Park, Hyun Ae Jung, Jin Won Kim, Han Jo Kim, Yongjun Cha, Sun Min Lim, Han Sang Kim, Choong-Kun Lee, Jee Hung Kim, Sang Hoon Chun, Jina Yun, So Yeon Park, Hye Seung Lee, Yong Mee Cho, Soo Jeong Nam, Kiyong Na, Sun Och Yoon, Ahwon Lee, Kee-Taek Jang, Hongseok Yun, Sungyoung Lee, Jee Hyun Kim, Wan-Seop Kim
- J Pathol Transl Med. 2024;58(4):147-164. Published online January 10, 2024
- DOI: https://doi.org/10.4132/jptm.2023.11.01
- 8,534 View
- 494 Download
- 1 Web of Science
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - In recent years, next-generation sequencing (NGS)–based genetic testing has become crucial in cancer care. While its primary objective is to identify actionable genetic alterations to guide treatment decisions, its scope has broadened to encompass aiding in pathological diagnosis and exploring resistance mechanisms. With the ongoing expansion in NGS application and reliance, a compelling necessity arises for expert consensus on its application in solid cancers. To address this demand, the forthcoming recommendations not only provide pragmatic guidance for the clinical use of NGS but also systematically classify actionable genes based on specific cancer types. Additionally, these recommendations will incorporate expert perspectives on crucial biomarkers, ensuring informed decisions regarding circulating tumor DNA panel testing.
-
Citations
Citations to this article as recorded by- Apport de la génomique dans la prise en charge des cancers
Étienne Rouleau, Lucie Karayan-Tapon, Marie-Dominique Galibert, Alexandre Harlé, Isabelle Soubeyran
Revue Francophone des Laboratoires.2025; 2025(568): 67. CrossRef - The Redox–Adhesion–Exosome (RAX) Hub in Cancer: Lipid Peroxidation-Driven EMT Plasticity and Ferroptosis Defense with HNE/MDA Signaling and Lipidomic Perspectives
Moon Nyeo Park, Jinwon Choi, Rosy Iara Maciel de Azambuja Ribeiro, Domenico V. Delfino, Seong-Gyu Ko, Bonglee Kim
Antioxidants.2025; 14(12): 1474. CrossRef
- Apport de la génomique dans la prise en charge des cancers
- Perspectives on single-nucleus RNA sequencing in different cell types and tissues
- Nayoung Kim, Huiram Kang, Areum Jo, Seung-Ah Yoo, Hae-Ock Lee
- J Pathol Transl Med. 2023;57(1):52-59. Published online January 10, 2023
- DOI: https://doi.org/10.4132/jptm.2022.12.19
- 23,083 View
- 472 Download
- 40 Web of Science
- 37 Crossref
-
 Abstract
Abstract
 PDF
PDF - Single-cell RNA sequencing has become a powerful and essential tool for delineating cellular diversity in normal tissues and alterations in disease states. For certain cell types and conditions, there are difficulties in isolating intact cells for transcriptome profiling due to their fragility, large size, tight interconnections, and other factors. Single-nucleus RNA sequencing (snRNA-seq) is an alternative or complementary approach for cells that are difficult to isolate. In this review, we will provide an overview of the experimental and analysis steps of snRNA-seq to understand the methods and characteristics of general and tissue-specific snRNA-seq data. Knowing the advantages and limitations of snRNA-seq will increase its use and improve the biological interpretation of the data generated using this technique.
-
Citations
Citations to this article as recorded by- Integrative Genomics Approach Identifies Glial Transcriptomic Dysregulation and Risk in the Cortex of Individuals With Alcohol Use Disorder
Anna S. Warden, Nihal A. Salem, Eric Brenner, Greg T. Sutherland, Julia Stevens, Manav Kapoor, Alison M. Goate, R. Dayne Mayfield
Biological Psychiatry.2026; 99(1): 34. CrossRef - Müller cell glutamine metabolism links photoreceptor and endothelial injury in diabetic retinopathy
Katia Corano Scheri, Yi-Wen Hsieh, Thomas Tedeschi, James B Hurley, Amani A Fawzi
Life Science Alliance.2026; 9(2): e202503434. CrossRef - Leveraging Single-Cell Technologies to Advance Understanding of Myocardial Disease
Robert S. Gardner, Nathan R. Tucker, Kaushik Amancherla
Circulation Research.2026;[Epub] CrossRef - Single-cell and spatial omics: exploring hypothalamic heterogeneity
Muhammad Junaid, Eun Jeong Lee, Su Bin Lim
Neural Regeneration Research.2025; 20(6): 1525. CrossRef - Exploring the utility of snRNA-seq in profiling human bladder tissue: A comprehensive comparison with scRNA-seq
Briana Santo, Emily E. Fink, Alexandra E. Krylova, Yi-Chia Lin, Mohamed Eltemamy, Alvin Wee, Oliver Wessely, Byron H. Lee, Angela H. Ting
iScience.2025; 28(1): 111628. CrossRef - Applications and emerging challenges of single-cell RNA sequencing technology in tumor drug discovery
Lu Zhang, Yueying Yang, Jianjun Tan
Drug Discovery Today.2025; 30(2): 104290. CrossRef - Techniques and analytic workflow for spatial transcriptomics and its application to allergy and inflammation
Haihan Zhang, Matthew T. Patrick, Jingyu Zhao, Xintong Zhai, Jialin Liu, Zheng Li, Yiqian Gu, Joshua Welch, Xiang Zhou, Robert L. Modlin, Lam C. Tsoi, Johann E. Gudjonsson
Journal of Allergy and Clinical Immunology.2025; 155(3): 678. CrossRef - Single-cell RNA sequencing in autoimmune diseases: New insights and challenges
Jialing Huang, Yuelin Hu, Shuqing Wang, Yuefang Liu, Xin Sun, Xin Wang, Hongsong Yu
Pharmacology & Therapeutics.2025; 267: 108807. CrossRef - SGK1 drives hippocampal demyelination and diabetes-associated cognitive dysfunction in mice
Ziying Jiang, Bin Liu, Tangsheng Lu, Xiaoxing Liu, Renjun Lv, Kai Yuan, Mengna Zhu, Xinning Wang, Shangbin Li, Song Xu, Xinyu Wang, Yifei Wang, Zhenfang Gao, Peiqing Zhao, Zongyong Zhang, Junwei Hao, Lin Lu, Qingqing Yin
Nature Communications.2025;[Epub] CrossRef - Unraveling cell–cell communication with NicheNet by inferring active ligands from transcriptomics data
Chananchida Sang-aram, Robin Browaeys, Ruth Seurinck, Yvan Saeys
Nature Protocols.2025; 20(6): 1439. CrossRef - A versatile and efficient method to isolate nuclei from low-input cryopreserved tissues for single-nuclei transcriptomics
Cristopher Segovia, Vincent Desrosiers, Fatemeh Khadangi, Karine Robitaille, Victoria Saavedra Armero, Myreille D’Astous, Gabriel Khelifi, Alain Bergeron, Samer Hussein, Maxime Richer, Yohan Bossé, Yves Fradet, Vincent Fradet, Steve Bilodeau
Scientific Reports.2025;[Epub] CrossRef - Application of single-cell sequencing technology and its clinical implications in Parkinson’s disease and Alzheimer’s disease: a narrative review
Zhonghao Chen, Jack Shi, Longfei Li
Advanced Technology in Neuroscience.2025; 2(1): 9. CrossRef - SGK1 upregulation in GFAP+ neurons in the frontal association cortex protects against neuronal apoptosis after spinal cord injury
Anbiao Wu, Guang Yang, Genyu Liu, Jiyan Zhang
Cell Death & Disease.2025;[Epub] CrossRef - Expert recommendations to standardize transcriptomic analysis in inflammatory bowel disease clinical trials
Bryan Linggi, Salas Azucena, Boyd Steere, Bram Verstockt, Dahham Alsoud, David Casero, Dermot McGovern, Eileen Chan, Michelle I Smith, Federica Ungaro, Florian Rieder, Konrad Aden, Lisa M Shackelton, Luca Massimino, Markus Neurath, Matthieu Allez, Raja At
Journal of Crohn's and Colitis.2025;[Epub] CrossRef - Transcriptional characterization of sepsis in a LPS porcine model
Ryan Neill
Molecular Genetics and Genomics.2025;[Epub] CrossRef - Single nuclear‐spatial transcriptomic sequencing reveals distinct puncture‐induced cell subpopulations in the intervertebral disc of a rat model
Guoyan Liang, Jing Tan, Chong Chen, Yuying Liu, Yongyu Ye, Xiaolin Pan, Qiujian Zheng, Yunbing Chang, Feng‐Juan Lyu
Clinical and Translational Medicine.2025;[Epub] CrossRef - Harp: data harmonization for computational tissue deconvolution across diverse transcriptomics platforms
Zahra Nozari, Paul Hüttl, Jakob Simeth, Marian Schön, James A Hutchinson, Rainer Spang, Macha Nikolski
Bioinformatics.2025;[Epub] CrossRef - Transformation of an Olfactory Placode-Derived Cell into One with Stem Cell Characteristics by Disrupting Epigenetic Barriers
Ghazia Abbas, Rutesh Vyas, Joyce C. Noble, Brian Lin, Robert P. Lane
Cellular Reprogramming.2025; 27(4): 164. CrossRef - Altered Neuroinflammatory Transcriptomic Profile in the Hippocampal Dentate Gyrus Three Weeks After Lateral Fluid Percussion Injury in Rats
Anthony J. DeSana, Yara Alfawares, Roshni Khatri, Tracy M. Hopkins, Faith V. Best, Jennifer L. McGuire, Laura B. Ngwenya
International Journal of Molecular Sciences.2025; 26(18): 9140. CrossRef - Methodologies for Sample Multiplexing and Computational Deconvolution in Single‐Cell Sequencing
Yufei Gao, Weiwei Yin, Wei Hu, Wei Chen
Advanced Science.2025;[Epub] CrossRef - A single-nucleus transcriptomic atlas of the adult Aedes aegypti mosquito
Olivia V. Goldman, Alexandra E. DeFoe, Yanyan Qi, Yaoyu Jiao, Shih-Che Weng, Brittney Wick, Leah Houri-Zeevi, Priyanka Lakhiani, Takeshi Morita, Jacopo Razzauti, Adriana Rosas-Villegas, Yael N. Tsitohay, Madison M. Walker, Ben R. Hopkins, Joshua X.D. Ang,
Cell.2025; 188(25): 7267. CrossRef - Leveraging single-cell RNA-seq in helminthology
Yi Mu, Chika P. Zumuk, Malcolm K. Jones, Pengfei Cai
Trends in Parasitology.2025;[Epub] CrossRef - Administration of a barcoded AAV capsid library to the putamen of non-human primates identifies variants with efficient retrograde transport
Yulia Dzhashiashvili, Jodi L. McBride, Emily Fabyanic, Xin Huang, Brian M. Kelly, Greglynn D. Walton-Gibbs, Vimala Vemireddi, Joan Wicks, Mohamad Nayal, Ariel A. Hippen, Zhenming Yu, Pichai Raman, Elizabeth Ramsburg, Marcus Davidsson, Esteban A. Engel, To
Molecular Therapy.2025;[Epub] CrossRef - Single-nucleus RNA sequencing resolves microenvironmental dynamics in brown/beige adipose tissue after bariatric surgery
Wei Wang, Yangxingyun Wang, Zhonghao Guo, Yao Lu, Wei Xie, Ruibin Li
Journal of Translational Medicine.2025;[Epub] CrossRef - Mapping the cellular landscape of Atlantic salmon head kidney by single cell and single nucleus transcriptomics
Adriana M.S. Andresen, Richard S. Taylor, Unni Grimholt, Rose Ruiz Daniels, Jianxuan Sun, Ross Dobie, Neil C. Henderson, Samuel A.M. Martin, Daniel J. Macqueen, Johanna H. Fosse
Fish & Shellfish Immunology.2024; 146: 109357. CrossRef - Single-cell and spatially resolved transcriptomics for liver biology
Ping Lin, Xi Yan, Siyu Jing, Yanhong Wu, Yiran Shan, Wenbo Guo, Jin Gu, Yu Li, Haibing Zhang, Hong Li
Hepatology.2024; 80(3): 698. CrossRef - Single-cell transcriptomics in thyroid eye disease
Sofia Ahsanuddin, Albert Y. Wu
Taiwan Journal of Ophthalmology.2024; 14(4): 554. CrossRef - Impaired cortical neuronal homeostasis and cognition after diffuse traumatic brain injury are dependent on microglia and type I interferon responses
Jonathan M. Packer, Chelsea E. Bray, Nicolas B. Beckman, Lynde M. Wangler, Amara C. Davis, Ethan J. Goodman, Nathaniel E. Klingele, Jonathan P. Godbout
Glia.2024; 72(2): 300. CrossRef - Adipose tissue macrophage heterogeneity in the single-cell genomics era
Haneul Kang, Jongsoon Lee
Molecules and Cells.2024; 47(2): 100031. CrossRef - A Comprehensive Review on Circulating cfRNA in Plasma: Implications for Disease Diagnosis and Beyond
Pengqiang Zhong, Lu Bai, Mengzhi Hong, Juan Ouyang, Ruizhi Wang, Xiaoli Zhang, Peisong Chen
Diagnostics.2024; 14(10): 1045. CrossRef - Single-Cell Sequencing Technology in Ruminant Livestock: Challenges and Opportunities
Avery Lyons, Jocelynn Brown, Kimberly M. Davenport
Current Issues in Molecular Biology.2024; 46(6): 5291. CrossRef - Single-Cell Transcriptomics Sheds Light on Tumor Evolution: Perspectives from City of Hope’s Clinical Trial Teams
Patrick A. Cosgrove, Andrea H. Bild, Thanh H. Dellinger, Behnam Badie, Jana Portnow, Aritro Nath
Journal of Clinical Medicine.2024; 13(24): 7507. CrossRef - Integrated analysis of single-cell and bulk RNA-seq establishes a novel signature for prediction in gastric cancer
Fei Wen, Xin Guan, Hai-Xia Qu, Xiang-Jun Jiang
World Journal of Gastrointestinal Oncology.2023; 15(7): 1215. CrossRef - Placental single cell transcriptomics: Opportunities for endocrine disrupting chemical toxicology
Elana R. Elkin, Kyle A. Campbell, Samantha Lapehn, Sean M. Harris, Vasantha Padmanabhan, Kelly M. Bakulski, Alison G. Paquette
Molecular and Cellular Endocrinology.2023; 578: 112066. CrossRef - Analyzing alternative splicing in Alzheimer’s disease postmortem brain: a cell-level perspective
Mohammad-Erfan Farhadieh, Kamran Ghaedi
Frontiers in Molecular Neuroscience.2023;[Epub] CrossRef - Single-nucleus transcriptome inventory of giant panda reveals cellular basis for fitness optimization under low metabolism
Shangchen Yang, Tianming Lan, Rongping Wei, Ling Zhang, Lin Lin, Hanyu Du, Yunting Huang, Guiquan Zhang, Shan Huang, Minhui Shi, Chengdong Wang, Qing Wang, Rengui Li, Lei Han, Dan Tang, Haimeng Li, Hemin Zhang, Jie Cui, Haorong Lu, Jinrong Huang, Yonglun
BMC Biology.2023;[Epub] CrossRef - Progress in research on tumor microenvironment-based spatial omics technologies
FANGMEI XIE, NAITE XI, ZEPING HAN, WENFENG LUO, JIAN SHEN, JINGGENG LUO, XINGKUI TANG, TING PANG, YUBING LV, JIABING LIANG, LIYIN LIAO, HAOYU ZHANG, YONG JIANG, YUGUANG LI, JINHUA HE
Oncology Research.2023; 31(6): 877. CrossRef
- Integrative Genomics Approach Identifies Glial Transcriptomic Dysregulation and Risk in the Cortex of Individuals With Alcohol Use Disorder
- Single-cell and spatial sequencing application in pathology
- Yoon-Seob Kim, Jinyong Choi, Sug Hyung Lee
- J Pathol Transl Med. 2023;57(1):43-51. Published online January 10, 2023
- DOI: https://doi.org/10.4132/jptm.2022.12.12
- 10,136 View
- 396 Download
- 10 Web of Science
- 12 Crossref
-
 Abstract
Abstract
 PDF
PDF - Traditionally, diagnostic pathology uses histology representing structural alterations in a disease’s cells and tissues. In many cases, however, it is supplemented by other morphology-based methods such as immunohistochemistry and fluorescent in situ hybridization. Single-cell RNA sequencing (scRNA-seq) is one of the strategies that may help tackle the heterogeneous cells in a disease, but it does not usually provide histologic information. Spatial sequencing is designed to assign cell types, subtypes, or states according to the mRNA expression on a histological section by RNA sequencing. It can provide mRNA expressions not only of diseased cells, such as cancer cells but also of stromal cells, such as immune cells, fibroblasts, and vascular cells. In this review, we studied current methods of spatial transcriptome sequencing based on their technical backgrounds, tissue preparation, and analytic procedures. With the pathology examples, useful recommendations for pathologists who are just getting started to use spatial sequencing analysis in research are provided here. In addition, leveraging spatial sequencing by integration with scRNA-seq is reviewed. With the advantages of simultaneous histologic and single-cell information, spatial sequencing may give a molecular basis for pathological diagnosis, improve our understanding of diseases, and have potential clinical applications in prognostics and diagnostic pathology.
-
Citations
Citations to this article as recorded by- Trends and Challenges of the Modern Pathology Laboratory for Biopharmaceutical Research Excellence
Sílvia Sisó, Anoop Murthy Kavirayani, Suzana Couto, Birgit Stierstorfer, Sunish Mohanan, Caroline Morel, Mathiew Marella, Dinesh S. Bangari, Elizabeth Clark, Annette Schwartz, Vinicius Carreira
Toxicologic Pathology.2025; 53(1): 5. CrossRef - Single-cell RNA sequencing in osteosarcoma: applications in diagnosis, prognosis, and treatment
Christèle Asmar, Guy Awad, Marc Boutros, Simon Daccache, Alain Chebly, Catherine Alix-Panabières, Hampig-Raphael Kourié
Medical Oncology.2025;[Epub] CrossRef - Characterizing Stroke Clots Using Single‐Cell Sequencing
Daniela Renedo, Tanyeri Barak, Jonathan DeLong, Julian N. Acosta, Nanthiya Sujijantarat, Andrew Koo, Cyprien A. Rivier, Santiago Clocchiatti‐Tuozzo, Shufan Huo, Joseph Antonios, James Giles, Guido J. Falcone, Kevin N. Sheth, Ryan Hebert, Murat Gunel, Laur
Journal of the American Heart Association.2025;[Epub] CrossRef - Spatial transcriptomics meets diabetic kidney disease: Illuminating the path to precision medicine
Dan-Dan Liu, Han-Yue Hu, Fei-Fei Li, Qiu-Yue Hu, Ming-Wei Liu, You-Jin Hao, Bo Li
World Journal of Diabetes.2025;[Epub] CrossRef - Spatial Transcriptomics in Pancreatic Cancer: Current Insights and Future Directions
Jin Su Kim
Journal of Digestive Cancer Research.2025; 13(3): 288. CrossRef - Incorporating Novel Technologies in Precision Oncology for Colorectal Cancer: Advancing Personalized Medicine
Pankaj Ahluwalia, Kalyani Ballur, Tiffanie Leeman, Ashutosh Vashisht, Harmanpreet Singh, Nivin Omar, Ashis K. Mondal, Kumar Vaibhav, Babak Baban, Ravindra Kolhe
Cancers.2024; 16(3): 480. CrossRef - Potential therapeutic targets for hypotension in duchenne muscular dystrophy
Harshi Saxena, Neal L. Weintraub, Yaoliang Tang
Medical Hypotheses.2024; 185: 111318. CrossRef - The crosstalk role of CDKN2A between tumor progression and cuproptosis resistance in colorectal cancer
Xifu Cheng, Famin Yang, Yuanheng Li, Yuke Cao, Meng Zhang, Jiameng JI, Yuxiao Bai, Qing Li, Qiongfang Yu, Dian Gao
Aging.2024; 16(12): 10512. CrossRef - Enquête exclusive sur le psoriasis
Imrane Ben Moussa, Bienfait Abasi-Ali, Fatima-Zahra Afarhkhane, Inès Mountadir, Claire Deligne
médecine/sciences.2024; 40(6-7): 584. CrossRef - Mechanisms of radiation‐induced tissue damage and response
Lin Zhou, Jiaojiao Zhu, Yuhao Liu, Ping‐Kun Zhou, Yongqing Gu
MedComm.2024;[Epub] CrossRef - A comparative analysis of single-cell transcriptomic technologies in plants and animals
Vamsidhar Reddy Netla, Harshraj Shinde, Gulshan Kumar, Ambika Dudhate, Jong Chan Hong, Ulhas Sopanrao Kadam
Current Plant Biology.2023; 35-36: 100289. CrossRef - Fibroblasts – the cellular choreographers of wound healing
Samuel Knoedler, Sonja Broichhausen, Ruiji Guo, Ruoxuan Dai, Leonard Knoedler, Martin Kauke-Navarro, Fortunay Diatta, Bohdan Pomahac, Hans-Guenther Machens, Dongsheng Jiang, Yuval Rinkevich
Frontiers in Immunology.2023;[Epub] CrossRef
- Trends and Challenges of the Modern Pathology Laboratory for Biopharmaceutical Research Excellence
- Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
- Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
- J Pathol Transl Med. 2022;56(5):249-259. Published online September 13, 2022
- DOI: https://doi.org/10.4132/jptm.2022.06.11
- 8,754 View
- 144 Download
- 7 Web of Science
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Activating mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) are predictive biomarkers for response to EGFR–tyrosine kinase inhibitor (TKI) therapy in lung adenocarcinoma (LUAD). Here, we characterized the clinicopathologic features associated with EGFR mutations via peptide nucleic acid clamping-assisted fluorescence melting curve analysis (PANAMutyper) and evaluated the feasibility of targeted deep sequencing for detecting the mutations.
Methods
We examined EGFR mutations in exons 18 through 21 for 2,088 LUADs from July 2017 to April 2020 using PANAMutyper. Of these, we performed targeted deep sequencing in 73 patients and evaluated EGFR-mutation status and TKI clinical response.
Results
EGFR mutation was identified in 55.7% of LUADs by PANAMutyper, with mutation rates higher in females (69.3%) and never smokers (67.1%) and highest in the age range of 50 to 59 years (64.9%). For the 73 patients evaluated using both methods, next-generation sequencing (NGS) identified EGFR mutation–positive results in 14 of 61 patients (23.0%) who were EGFR-negative according to PANAMutyper testing. Of the 10 patients reportedly harboring a sensitizing mutation according to NGS, seven received TKI treatment, with all showing partial response or stable disease. In the 12 PANAMutyper-positive cases, NGS identified two additional mutations in exon 18, whereas a discordant negative result was observed in two cases.
Conclusions
Although PANAMutyper identified high frequencies of EGFR mutations, targeted deep sequencing revealed additional uncommon EGFR mutations. These findings suggested that appropriate use of NGS may benefit LUAD patients with otherwise negative screening test results. -
Citations
Citations to this article as recorded by- Comparison of tissue-based and plasma-based testing for EGFR mutation in non–small cell lung cancer patients
Yoon Kyung Kang, Dong Hoon Shin, Joon Young Park, Chung Su Hwang, Hyun Jung Lee, Jung Hee Lee, Jee Yeon Kim, JooYoung Na
Journal of Pathology and Translational Medicine.2025; 59(1): 60. CrossRef - Localization of epidermal growth factor receptor-mutations using PNA:DNA probes in clinical specimens from patients with non-small cell lung cancer
Haruo Miyata, Hajime Shigeto, Tomoatsu Ikeya, Tadashi Ashizawa, Akira Iizuka, Yasufumi Kikuchi, Chie Maeda, Akari Kanematsu, Kazue Yamashita, Kenichi Urakami, Yuji Shimoda, Takeshi Nagashima, Keiichi Ohshima, Yasuhisa Ohde, Mitsuhiro Isaka, Takashi Sugino
Scientific Reports.2025;[Epub] CrossRef - Molecular characteristics and responses to EGFR tyrosine kinase inhibitors in non-small cell lung cancer patients with EGFR exon 19 insertions
Yang Li, Yunfeng Ni, Feng Lv, Yan Shi, Yedan Chen, Xiaoying Wu, Jiaohui Pang, Long Huang, Yang Shao, Tao Wang, Jie Min, Yang Song
BMC Medicine.2025;[Epub] CrossRef - Detection of EGFR exon 20 insertion mutations in non-small cell lung cancer: implications for consistent nomenclature in precision medicine
Jieun Park, Boram Lee, Ji-Young Song, Minjung Sung, Mi Jeong Kwon, Chae Rin Kim, Sangjin Lee, Young Kee Shin, Yoon-La Choi
Pathology.2024; 56(5): 653. CrossRef - Histo-pillar strip for optimal histogel block construction and biomarker analysis in 3D-lung cancer patient-derived organoids
Sang-Yun Lee, Eunyoung Lee, Ji-O Ryu, Kyuhwan Kim, Yongki Hwang, Bosung Ku, Seok Whan Moon, Mi Hyoung Moon, Kyung Soo Kim, Kwanyong Hyun, Jeong Uk Lim, Chan Kwon Park, Sung Won Kim, Chang Dong Yeo, Dong Woo Lee, Seung Joon Kim
Biofabrication.2024; 16(4): 045017. CrossRef
- Comparison of tissue-based and plasma-based testing for EGFR mutation in non–small cell lung cancer patients
- Lymphoproliferative disorder involving body fluid: diagnostic approaches and roles of ancillary studies
- Jiwon Koh, Sun Ah Shin, Ji Ae Lee, Yoon Kyung Jeon
- J Pathol Transl Med. 2022;56(4):173-186. Published online July 4, 2022
- DOI: https://doi.org/10.4132/jptm.2022.05.16
- 10,792 View
- 319 Download
- 4 Web of Science
- 4 Crossref
-
 Abstract
Abstract
 PDF
PDF - Lymphocyte-rich effusions represent benign reactive process or neoplastic condition. Involvement of lymphoproliferative disease in body cavity is not uncommon, and it often causes diagnostic challenge. In this review, we suggest a practical diagnostic approach toward lymphocyte-rich effusions, share representative cases, and discuss the utility of ancillary tests. Cytomorphologic features favoring neoplastic condition include high cellularity, cellular atypia/pleomorphism, monomorphic cell population, and frequent apoptosis, whereas lack of atypia, polymorphic cell population, and predominance of small T cells usually represent benign reactive process. Involvement of non-hematolymphoid malignant cells in body fluid should be ruled out first, followed by categorization of the samples into either small/medium-sized cell dominant or large-sized cell dominant fluid. Small/medium-sized cell dominant effusions require ancillary tests when either cellular atypia or history/clinical suspicion of lymphoproliferative disease is present. Large-sized cell dominant effusions usually suggest neoplastic condition, however, in the settings of initial presentation or low overall cellularity, ancillary studies are helpful for more clarification. Ancillary tests including immunocytochemistry, in situ hybridization, clonality test, and next-generation sequencing can be performed using cytologic preparations. Throughout the diagnostic process, proper review of clinical history, cytomorphologic examination, and application of adequate ancillary tests are key elements for successful diagnosis.
-
Citations
Citations to this article as recorded by- The case of the sneaky lymphoma: solved by flow cytometry
Renu Singh, Md Ali Osama, Rachana Meena, Shailaja Shukla, Jagdish Chandra
Indian Journal of Thoracic and Cardiovascular Surgery.2025; 41(9): 1258. CrossRef - The urgency of Burkitt lymphoma diagnosis in fluid cytology—A tertiary care experience
Soundarya Ravi, Anu K. Devi, Prabhu Manivannan, Debasis Gochhait, Rakhee Kar, Neelaiah Siddaraju
Cytopathology.2024; 35(2): 275. CrossRef - Immunocytochemistry on frozen-embedded cell block for the diagnosis of hematolymphoid cytology specimen: a straightforward alternative to the conventional cell block
Youjeong Seo, Sanzida Alam Prome, Lucia Kim, Jee Young Han, Joon Mee Kim, Suk Jin Choi
Journal of Hematopathology.2024; 17(1): 1. CrossRef - Lymphoma presenting as the first finding in pleural fluid cytology: A rare cytologic presentation
Kafil Akhtar, Gowthami Nagendhran, Anjum Ara, Masheera Akhtar
IP Archives of Cytology and Histopathology Research.2024; 8(4): 250. CrossRef
- The case of the sneaky lymphoma: solved by flow cytometry
- Adrenal hemangioblastoma
- Joo-Yeon Koo, Kyung-Hwa Lee, Joon Hyuk Choi, Ho Seok Chung, Chan Choi
- J Pathol Transl Med. 2022;56(3):161-166. Published online February 28, 2022
- DOI: https://doi.org/10.4132/jptm.2021.12.28
- 5,492 View
- 157 Download
- 1 Web of Science
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - Hemangioblastoma (HB) is a rare benign tumor that most commonly occurs in the cerebellum. HB is composed of neoplastic stromal cells and abundant small vessels. However, the exact origin of stromal cells is controversial. Extraneural HBs have been reported in a small series, and peripheral HBs arising in the adrenal gland are extremely rare. Herein, we report a case of sporadic adrenal HB in a 54-year-old woman. The tumor was a well-circumscribed, yellow mass measuring 4.2 cm in diameter. Histologically, the tumor was composed of small blood vessels and vacuolated stromal cells with clear cytoplasm. On immunohistochemical stain, the stromal cells were positive for S-100 protein, neuron-specific enolase, and synaptophysin. The tumor did not reveal mutation of VHL alleles. We herein present a case of HB of the adrenal gland and review of the literature.
-
Citations
Citations to this article as recorded by- Familial Von Hippel–Lindau Disease: A Case Series of Cerebral Hemangioblastomas with MRI, Histopathological, and Genetic Correlations
Claudiu Matei, Ioana Boeras, Dan Orga Dumitriu, Cosmin Mutu, Adriana Popescu, Mihai Gabriel Cucu, Alexandru Calotă-Dobrescu, Bogdan Fetica, Diter Atasie
Life.2025; 15(11): 1649. CrossRef
- Familial Von Hippel–Lindau Disease: A Case Series of Cerebral Hemangioblastomas with MRI, Histopathological, and Genetic Correlations
- An unusual case of microsatellite instability–high/deficient mismatch repair (MSI-H/dMMR) diffuse large B-cell lymphoma revealed by targeted gene sequencing
- Bogyeong Han, Sehui Kim, Jiwon Koh, Jeong Mo Bae, Hongseok Yun, Yoon Kyung Jeon
- J Pathol Transl Med. 2022;56(2):92-96. Published online November 16, 2021
- DOI: https://doi.org/10.4132/jptm.2021.10.15
- 9,124 View
- 263 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - Microsatellite instability-high/deficient mismatch repair (MSI-H/dMMR) status has been approved as a tissue-agnostic biomarker for immune checkpoint inhibitor therapy in patients with solid tumors. We report the case of an MSI-H/dMMR diffuse large B-cell lymphoma (DLBCL) identified by targeted gene sequencing (TGS). A 90-year-old female who presented with vaginal bleeding and a large mass in the upper vagina was diagnosed with germinal center-B-cell-like DLBCL, which recurred at the uterine cervix at 9 months after chemotherapy. Based on TGS of 121 lymphoma-related genes and the LymphGen algorithm, the tumor was classified genetically as DLBCL of EZB subtype. Mutations in multiple genes, including frequent frameshift mutations, were detected by TGS and further suggested MSI. The MSI-H/dMMR and loss of MLH1 and PMS2 expression were determined in MSI-fragment analysis, MSI real-time polymerase chain reaction, and immunohistochemical tests. This case demonstrates the potential diagnostic and therapeutic utility of lymphoma panel sequencing for DLBCL with MSI-H/dMMR.
-
Citations
Citations to this article as recorded by- Shared genomic features of HIV+ diffuse large B-cell lymphoma in two African cohorts
Sophia M. Roush, Mishalan Moodley, Jenny Coelho, Samantha Beck, Amon Chirwa, Edwards Kasonkanji, Marriam Mponda, Maurice Mulenga, Tamiwe Tomoka, Hanri van Zijl, Katherine Hodkinson, Arshad Ismail, Senzo Mtshali, Jonathan Featherston, Satish Gopal, Matthew
Scientific Reports.2025;[Epub] CrossRef - Chimeric and mutant CARD9 constructs enable analyses of conserved and diverged autoinhibition mechanisms in the CARD‐CC protein family
Jens Staal, Yasmine Driege, Femke Van Gaever, Jill Steels, Rudi Beyaert
The FEBS Journal.2024; 291(6): 1220. CrossRef - PD-L1+diffuse large B-cell lymphoma with extremely high mutational burden and microsatellite instability due to acquiredPMS2mutation
Andrew W. Allbee, James Gerson, Guang Yang, Adam Bagg
Molecular Case Studies.2023; 9(4): a006318. CrossRef
- Shared genomic features of HIV+ diffuse large B-cell lymphoma in two African cohorts
- A study of pathological characteristics and BRAF V600E status in Langerhans cell histiocytosis of Vietnamese children
- Thu Dang Anh Phan, Bao Gia Phung, Tu Thanh Duong, Vu Anh Hoang, Dat Quoc Ngo, Nguyen Dinh The Trinh, Tung Thanh Tran
- J Pathol Transl Med. 2021;55(2):112-117. Published online January 27, 2021
- DOI: https://doi.org/10.4132/jptm.2020.11.30
- 7,074 View
- 129 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
Langerhans cell histiocytosis (LCH) is more common in children than adults and involves many organs. In children, the BRAF V600E mutation is associated with recurrent and high-risk LCH.
Methods
We collected paraffin blocks of 94 pediatric LCH patients to detect BRAF V600E mutation by sequencing. The relationship between BRAF V600E status and clinicopathological parameters were also critically analyzed.
Results
BRAF V600E mutation exon 15 was detected in 45 cases (47.9%). Multiple systems LCH showed a significantly higher BRAF V600E mutation rate than a single system (p=.001). No statistical significance was evident for other clinical characteristics such as age, sex, location, risk organs involvement, and CD1a expression.
Conclusions
In Vietnamese LCH children, the proportion of BRAF V600E mutational status was relatively high and related to multiple systems. -
Citations
Citations to this article as recorded by- Clinicopathological features of hepatic Langerhans cell histiocytosis: report of ten cases and review of the literature
Qian-Qian Chen, Chun-kui Shao, Yi-wang Zhang, Jian-ning Chen, Hai-feng Li, Qiong Liang
Annals of Diagnostic Pathology.2026; 81: 152586. CrossRef - Pathologic characteristics of histiocytic and dendritic cell neoplasms
Sun Och Yoon
Blood Research.2024;[Epub] CrossRef - Sulfur dots/Au@Ag nanorods array-based polarized ECL sensor for the detection of thyroid cancer biomarker
Zixuan Ding, Peilin Wang, Zhenrun Li, Yupeng Guo, Qiang Ma
Talanta.2023; 265: 124925. CrossRef
- Clinicopathological features of hepatic Langerhans cell histiocytosis: report of ten cases and review of the literature
- A case of concomitant EGFR/ALK alteration against a mutated EGFR background in early-stage lung adenocarcinoma
- Ki-Chang Lee, Jiwon Koh, Doo Hyun Chung, Yoon Kyung Jeon
- J Pathol Transl Med. 2021;55(2):139-144. Published online January 22, 2021
- DOI: https://doi.org/10.4132/jptm.2020.12.16
- 5,223 View
- 111 Download
- 4 Web of Science
- 4 Crossref
-
 Abstract
Abstract
 PDF
PDF - Rare cases of lung adenocarcinoma (LUAD) with concomitant epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) translocation have been reported. However, their clonal and evolutional relationship remains unclear. We report a case of early-stage EGFR-mutated LUAD with a focal concomitant EGFR/ALK alteration. A 63-year-old male underwent lobectomy to remove a 1.9-cm-sized lung nodule, which was diagnosed with EGFR-mutated LUAD. ALK immunohistochemistry (IHC) showed focal positivity within the part of the tumor characterized by lepidic pattern, also confirmed by fluorescence in-situ hybridization (FISH). Targeted next-generation sequencing was performed separately on the ALK IHC/FISH-positive and -negative areas. EGFR L833V/L858R mutations were detected in both areas, whereas EML4 (echinoderm microtubule-associated protein-like 4)-ALK translocations was confirmed only in the ALK IHC/FISH-positive area, suggesting the divergence of an EGFR/ALK co-altered subclone from the original EGFR-mutant clone. Our study suggests that concurrent alterations of EGFR and ALK can arise via divergent tumor evolution, even in the relatively early phases of tumorigenesis.
-
Citations
Citations to this article as recorded by- Machine learning-based characterization of a PANoptosis-associated model for enhancing prognosis and immunotherapy response in lung adenocarcinoma patients
Ziqiao Fu, Jia Zeng, Xiaomin Xiong, Weimin Zhong
Discover Oncology.2025;[Epub] CrossRef - Identification and validation of molecular subtype and prognostic signature for lung adenocarcinoma based on neutrophil extracellular traps
Yanhua Zuo, Guangyi Leng, Ping Leng
Pathology and Oncology Research.2023;[Epub] CrossRef - Machine Learning-Based Integration Develops a Macrophage-Related Index for Predicting Prognosis and Immunotherapy Response in Lung Adenocarcinoma
Zuwei Li, Minzhang Guo, Wanli Lin, Peiyuan Huang
Archives of Medical Research.2023; 54(7): 102897. CrossRef - Big data analysis identified a telomere-related signature predicting the prognosis and drug sensitivity in lung adenocarcinoma
Weiyi Zhang
Medicine.2023; 102(46): e35526. CrossRef
- Machine learning-based characterization of a PANoptosis-associated model for enhancing prognosis and immunotherapy response in lung adenocarcinoma patients
- DNA-protein biomarkers for immunotherapy in the era of precision oncology
- Binnari Kim, So Young Kang, Kyoung-Mee Kim
- J Pathol Transl Med. 2021;55(1):26-32. Published online November 9, 2020
- DOI: https://doi.org/10.4132/jptm.2020.09.23
- 6,912 View
- 186 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - The use of biomarkers to guide patient and therapy selection has gained much attention to increase the scope and complexity of targeted therapy options and immunotherapy. Clinical trials provide a basis for discovery of biomarkers, which can then aid in development of new drugs. To that end, samples from cancer patients, including DNA, RNA, protein, and the metabolome isolated from cancer tissues and blood or urine, are analyzed in various ways to identify relevant biomarkers. In conjunction with nucleotide-based, high-throughput, next-generation sequencing techniques, therapy-guided biomarker assays relying on protein-based immunohistochemistry play a pivotal role in cancer care. In this review, we discuss the current knowledge regarding DNA and protein biomarkers for cancer immunotherapy
-
Citations
Citations to this article as recorded by- Treatment Selection for Patients with HER2-Negative Metastatic Gastric Cancer Expressing Claudin 18.2 and PD-L1
Yusuke Miyajima, Takeshi Kawakami
Cancers.2025; 17(7): 1120. CrossRef - NCKAP1 as a prognostic and immunological biomarker: pan-cancer analysis and validation in renal clear cell carcinoma
Xiao Liang
American Journal of Translational Research.2024; 16(8): 4083. CrossRef - Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer
Moonsik Kim, Ji Yun Jeong, An Na Seo
Diagnostics.2023; 13(17): 2782. CrossRef
- Treatment Selection for Patients with HER2-Negative Metastatic Gastric Cancer Expressing Claudin 18.2 and PD-L1
- Prediction of TP53 mutations by p53 immunohistochemistry and their prognostic significance in gastric cancer
- Hye Jung Hwang, Soo Kyung Nam, Hyunjin Park, Yujun Park, Jiwon Koh, Hee Young Na, Yoonjin Kwak, Woo Ho Kim, Hye Seung Lee
- J Pathol Transl Med. 2020;54(5):378-386. Published online July 1, 2020
- DOI: https://doi.org/10.4132/jptm.2020.06.01
- 12,858 View
- 287 Download
- 41 Web of Science
- 37 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Recently, molecular classifications of gastric cancer (GC) have been proposed that include TP53 mutations and their functional activity. We aimed to demonstrate the correlation between p53 immunohistochemistry (IHC) and TP53 mutations as well as their clinicopathological significance in GC.
Methods
Deep targeted sequencing was performed using surgical or biopsy specimens from 120 patients with GC. IHC for p53 was performed and interpreted as strong, weak, or negative expression. In 18 cases (15.0%) with discrepant TP53 mutation and p53 IHC results, p53 IHC was repeated.
Results
Strong expression of p53 was associated with TP53 missense mutations, negative expression with other types of mutations, and weak expression with wild-type TP53 (p<.001). The sensitivity for each category was 90.9%, 79.0%, and 80.9%, and the specificity was 95.4%, 88.1%, and 92.3%, respectively. The TNM stage at initial diagnosis exhibited a significant correlation with both TP53 mutation type (p=.004) and p53 expression status (p=.029). The Kaplan-Meier survival analysis for 109 stage II and III GC cases showed that patients with TP53 missense mutations had worse overall survival than those in the wild-type and other mutation groups (p=.028). Strong expression of p53 was also associated with worse overall survival in comparison to negative and weak expression (p=.035).
Conclusions
Results of IHC of the p53 protein may be used as a simple surrogate marker of TP53 mutations. However, negative expression of p53 and other types of mutations of TP53 should be carefully interpreted because of its lower sensitivity and different prognostic implications. -
Citations
Citations to this article as recorded by- The future is now: advancing p53 immunohistochemistry in Barrett's oesophagus and its implication for the everyday pathologist
Yevgen Chornenkyy, Monika Vyas, Vikram Deshpande
Histopathology.2026; 88(2): 380. CrossRef - Advancement in preclinical development of cancer treatment agents through modulation of Rac1: From EHop-016 to natural products
Yingyi Liu, Sze-Nga Wong, Aiping Lyu, Joshua Ka-Shun Ko
Biochimica et Biophysica Acta (BBA) - Reviews on Cancer.2026; 1881(1): 189522. CrossRef - Tumor-Associated Macrophage Infiltration and PD-L1 Expression in Gastric Cancer According to a Modified TCGA-Based Classification
Boram Song, Dong-Hoe Koo, Eo Jin Kim, In-Gu Do, Jinah Chu, Kyungeun Kim, Hyebin Lee, Min-Jung Kwon, Jung Ho Park, Byung Ho Son, Chang Hak Yoo, Seoung Wan Chae
Journal of Gastric Cancer.2026;[Epub] CrossRef - Linking p53 immunostaining to TP53 mutation status in patients with non-small cell lung cancer
Taeyeong Kim, Suyeon Kim, Sangjin Lee, Soohyun Hwang, Joungho Han, Hoyeon Jeong, Yoon-La Choi
Pathology.2025; 57(7): 881. CrossRef - Correlation of TP53 Genetic Alterations with p53 Immunohistochemical Expression and Their Prognostic Significance in DLBCL
Chen Chen, Zijuan Hu, Min Ren, Longlong Bao, Ran Wei, Tian Tian, Xiaoli Zhu, Qianming Bai, Baohua Yu, Xiaoqiu Li, Xiaoyan Zhou
Current Oncology.2025; 32(9): 488. CrossRef - Immunophenotypic Panel for Comprehensive Characterization of Aggressive Thyroid Carcinomas
Mihail Ceausu, Mihai Alin Publik, Dana Terzea, Carmen Adina Cristea, Dumitru Ioachim, Dana Manda, Sorina Schipor
Cells.2025; 14(19): 1554. CrossRef - Multiple approaches revealed MGc80‐3 as a somatic hybrid with HeLa cells rather than a gastric cancer cell line
Fang Cao, Hao Sun, Zhenli Yang, Yanhua Bai, Xiao Hu, Yuhong Hou, Xiaocui Bian, Yuqin Liu
International Journal of Cancer.2024; 154(1): 155. CrossRef - In Response to p53 Immunohistochemical Staining and TP53 Gene Mutations in Endometrial Cancer: Does Null Pattern Correlate With Prognosis?
Ikuko Sakamoto, Keiko Kagami, Takahiro Nozaki, Yosuke Hirotsu, Kenji Amemiya, Toshio Oyama, Masao Omata
American Journal of Surgical Pathology.2024; 48(3): 374. CrossRef - CHEK2 germline variants identified in familial nonmedullary thyroid cancer lead to impaired protein structure and function
Carolina Pires, Inês J. Marques, Mariana Valério, Ana Saramago, Paulo E. Santo, Sandra Santos, Margarida Silva, Margarida M. Moura, João Matos, Teresa Pereira, Rafael Cabrera, Diana Lousa, Valeriano Leite, Tiago M. Bandeiras, João B. Vicente, Branca M. Ca
Journal of Biological Chemistry.2024; 300(3): 105767. CrossRef - The spectrum of TP53 mutations in Rwandan patients with gastric cancer
Augustin Nzitakera, Jean Bosco Surwumwe, Ella Larissa Ndoricyimpaye, Schifra Uwamungu, Delphine Uwamariya, Felix Manirakiza, Marie Claire Ndayisaba, Gervais Ntakirutimana, Benoit Seminega, Vincent Dusabejambo, Eric Rutaganda, Placide Kamali, François Ngab
Genes and Environment.2024;[Epub] CrossRef - Gastric cancer molecular classification based on immunohistochemistry and in‐situ hybridisation and mortality
Maarit Eskuri, Eva‐Maria Birkman, Joonas H Kauppila
Histopathology.2024; 85(2): 327. CrossRef - Redefining aberrant P53 expression of gastric cancer and its distinct clinical significance among molecular-histologic subtypes
Shih-Chiang Huang, Ian Yi-Feng Chang, Tse-Ching Chen, Hsiao-Ching Lin, Chun-Yi Tsai, Jun-Te Hsu, Chun-Nan Yeh, Shih-Cheng Chang, Ta-Sen Yeh
Asian Journal of Surgery.2024; 47(11): 4699. CrossRef - Assessment of TP53 and CDKN2A status as predictive markers of malignant transformation of sinonasal inverted papilloma
Soohyeon Kwon, Jeong-Whun Kim, Eun Sun Kim, Jin Ho Paik, Jin-Haeng Chung, Sung-Woo Cho, Tae-Bin Won, Chae-Seo Rhee, Jee Hye Wee, Hyojin Kim
Scientific Reports.2024;[Epub] CrossRef - Implementing an integrated molecular classification for gastric cancer from endoscopic biopsies using on-slide tests
Simona Costache, Adelina Baltan , Sofia Diaz McLinn , Mattia Pegoraro , Rebecca de Havilland , Matthew Porter , Ana Lerga , Teresa Thomas , Alina Elena Chefani
Romanian Journal of Morphology and Embryology.2024; 65(2): 257. CrossRef - Application of NGS molecular classification in the diagnosis of endometrial carcinoma: A supplement to traditional pathological diagnosis
Qunxian Rao, Jianwei Liao, Yangyang Li, Xin Zhang, Guocai Xu, Changbin Zhu, Shengya Tian, Qiuhong Chen, Hui Zhou, Bingzhong Zhang
Cancer Medicine.2023; 12(5): 5409. CrossRef - Predictive value of p53 and AXL immunostaining for the efficacy of immune checkpoint inhibitor-based therapy after osimertinib treatment in patients with epidermal growth factor-mutant non-small cell lung cancer
Kenji Morimoto, Tadaaki Yamada, Ryo Sawada, Koichi Azuma, Yasuhiro Goto, Taishi Harada, Shinsuke Shiotsu, Nobuyo Tamiya, Yusuke Chihara, Takayuki Takeda, Osamu Hiranuma, Isao Hasegawa, Satomi Tanaka, Akihiro Yoshimura, Masahiro Iwasaku, Shinsaku Tokuda, Y
Cancer Immunology, Immunotherapy.2023; 72(6): 1699. CrossRef - Validation of p53 Immunohistochemistry (PAb240 Clone) in Canine Tumors with Next-Generation Sequencing (NGS) Analysis
Barbara Brunetti, Dario de Biase, Giulia Dellapina, Luisa Vera Muscatello, Francesco Ingravalle, Giorgia Tura, Barbara Bacci
Animals.2023; 13(5): 899. CrossRef - Mesonephric‐like adenocarcinoma of the female genital tract: novel observations and detailed molecular characterisation of mixed tumours and mesonephric‐like carcinosarcomas
Jelena Mirkovic, Ekaterina Olkhov‐Mitsel, Yutaka Amemiya, Maysa Al‐Hussaini, Sharon Nofech‐Mozes, Bojana Djordjevic, Rachel Kupets, Arun Seth, W Glenn McCluggage
Histopathology.2023; 82(7): 978. CrossRef - Clinicopathologic characterization of cervical metastasis from an unknown primary tumor: a multicenter study in Korea
Miseon Lee, Uiree Jo, Joon Seon Song, Youn Soo Lee, Chang Gok Woo, Dong-Hoon Kim, Jung Yeon Kim, Sun Och Yoon, Kyung-Ja Cho
Journal of Pathology and Translational Medicine.2023; 57(3): 166. CrossRef - P53 in Penile Squamous Cell Carcinoma: A Pattern-Based Immunohistochemical Framework with Molecular Correlation
Isabel Trias, Adela Saco, Lorena Marimon, Ricardo López del Campo, Carolina Manzotti, Oriol Ordi, Marta del Pino, Francisco M. Pérez, Naiara Vega, Silvia Alós, Antonio Martínez, Leonardo Rodriguez-Carunchio, Oscar Reig, Pedro Jares, Cristina Teixido, Tare
Cancers.2023; 15(10): 2719. CrossRef - p53/TP53 Status Assessment in Gastroesophageal Adenocarcinoma
Elisa Boldrin, Maria Assunta Piano, Francesco Bernaudo, Rita Alfieri, Maria Raffaella Biasin, Isabella Monia Montagner, Alice Volpato, Genny Mattara, Francesco Lamacchia, Giovanna Magni, Antonio Rosato, Antonio Scapinello, Pierluigi Pilati, Matteo Curtare
Cancers.2023; 15(10): 2783. CrossRef - Genomic profiling of dedifferentiated endometrial carcinomas arising in the background of high‐grade carcinoma: a targeted next‐generation sequencing study
Ekaterina Olkhov‐Mitsel, Aurelia Busca, Carlos Parra‐Herran, Yutaka Amemiya, Sharon Nofech‐Mozes, Bojana Djordjevic, Marisa R Nucci, Arun Seth, Jelena Mirkovic
Histopathology.2023; 83(3): 366. CrossRef -
Clinicopathologic Features and Prognostic Significance of Immunohistochemistry and In Situ Hybridization Based Molecular Classification in Gastric Carcinoma
Gizem Issin, İlyas Sayar, Fatih Demir, İrem Güvendir Bakkaloğlu, Mehmet Gamsizkan, Zeliha Yildiz, Ismail Yilmaz, Sevilay Akalp Özmen, Diren Vuslat Çağatay, Itır Ebru Zemheri, Murat Demiriz, Armağan Günal
Journal of Environmental Pathology, Toxicology and Oncology.2023; 42(4): 1. CrossRef - Clinicopathologic and Molecular Characterization of Anorectal Neuroendocrine Carcinomas Reveals Human Papillomavirus, p53, and c-Myc as Alternative Mechanisms of Carcinogenesis
Allison J. Cox, William E. Crowe, Qi Yang, Bin Zhang, Zoltán N. Oltvai, Xiaoyan Liao
Modern Pathology.2023; 36(11): 100295. CrossRef - Dedifferentiated Endometrial Carcinoma: A Rare Aggressive Neoplasm-Clinical, Morphological and Immunohistochemical Features
Giovanna Giordano, Elena Ferioli, Debora Guareschi, Alessandro Tafuni
Cancers.2023; 15(21): 5155. CrossRef - Characterization on the oncogenic effect of the missense mutations of p53 via machine learning
Qisheng Pan, Stephanie Portelli, Thanh Binh Nguyen, David B Ascher
Briefings in Bioinformatics.2023;[Epub] CrossRef - Adrenal Nodules Detected at Staging CT in Patients with Resectable Gastric Cancers Have a Low Incidence of Malignancy
Hae Young Kim, Won Chang, Yoon Jin Lee, Ji Hoon Park, Jungheum Cho, Hee Young Na, Hyungwoo Ahn, Sung Il Hwang, Hak Jong Lee, Young Hoon Kim, Kyoung Ho Lee
Radiology.2022; 302(1): 129. CrossRef - Intestinal-type gastric dysplasia in Helicobacter pylori-naïve patients
Kotaro Shibagaki, Ayako Itawaki, Yoichi Miyaoka, Kenichi Kishimoto, Yusuke Takahashi, Satoshi Kotani, Tsuyoshi Mishiro, Naoki Oshima, Kousaku Kawashima, Norihisa Ishimura, Hideyuki Onuma, Makoto Nagasaki, Mamiko Nagase, Asuka Araki, Kyuichi Kadota, Ryoji
Virchows Archiv.2022; 480(4): 783. CrossRef - Dedifferentiation-like tubular and solid carcinoma of the stomach shows phenotypic divergence and association with deficient SWI/SNF complex
Shih-Chiang Huang, Kuang-Hua Chen, Kwai-Fong Ng, I-Chieh Lin, Yi-Chun Chao, Ta-Sen Yeh, Huei-Chieh Chuang, Tse-Ching Chen
Virchows Archiv.2022; 480(4): 771. CrossRef - Distinct molecular phenotype and the potential prognostic value of immune prognostic index and tumor infiltrating lymphocytes in hepatoid adenocarcinoma of stomach
Muxing Kang, Xiaojing Ma, Jifei Shi, Guofeng Chen, Xiaoli Jin, Jun Wang, Lele Lin, Zhiwei Wu, Kaibo Chen, Jinghong Xu, Pintong Huang, Jian Chen
Translational Oncology.2022; 19: 101380. CrossRef - Evaluation of Tumor DNA Sequencing Results in Patients with Gastric and Gastroesophageal Junction Adenocarcinoma Stratified by TP53 Mutation Status
Anthony C Wood, Yonghong Zhang, Qianxing Mo, Ling Cen, Jacques Fontaine, Sarah E Hoffe, Jessica Frakes, Sean P Dineen, Jose M Pimiento, Christine M Walko, Rutika Mehta
The Oncologist.2022; 27(4): 307. CrossRef - Comprehensive Clinical Analysis of Gallbladder Neuroendocrine Neoplasms: A Large-Volume Multicenter Study During One Decade
Yangyang Wang, Bingfeng Huang, Qihan Fu, Jianing Wang, Mao Ye, Manyi Hu, Kai Qu, Kai Liu, Xiao Hu, Shumei Wei, Ke Sun, Wenbo Xiao, Bo Zhang, Haijun Li, Jingsong Li, Qi Zhang, Tingbo Liang
Annals of Surgical Oncology.2022; 29(12): 7619. CrossRef - Expression of SASP, DNA Damage Response, and Cell Proliferation Factors in Early Gastric Neoplastic Lesions: Correlations and Clinical Significance
Li Liang, Yijie Chai, Fei Chai, Haijing Liu, Ningning Ma, Hong Zhang, Shuang Zhang, Lin Nong, Ting Li, Bo Zhang
Pathology and Oncology Research.2022;[Epub] CrossRef - Systems biology and OMIC data integration to understand gastrointestinal cancers
Iasmin Moreira Costa Bispo, Henry Paul Granger, Palloma Porto Almeida, Patricia Belini Nishiyama, Leandro Martins de Freitas
World Journal of Clinical Oncology.2022; 13(10): 762. CrossRef - MicroRNA-552 expression in colorectal cancer and its clinicopathological significance
Joon Im, Soo Kyung Nam, Hye Seung Lee
Journal of Pathology and Translational Medicine.2021; 55(2): 125. CrossRef - Different effects of p53 protein overexpression on the survival of gastric cancer patients according to Lauren histologic classification: a retrospective study
Ki Wook Kim, Nayoung Kim, Yonghoon Choi, Won Seok Kim, Hyuk Yoon, Cheol Min Shin, Young Soo Park, Dong Ho Lee, Young Suk Park, Sang-Hoon Ahn, Do Joong Park, Hyung-Ho Kim, Hye Seung Lee, Ji-Won Kim, Jin Won Kim, Keun-Wook Lee, Won Chang, Ji Hoon Park, Yoon
Gastric Cancer.2021; 24(4): 844. CrossRef - The association between the expression of nuclear Yes-associated protein 1 (YAP1) and p53 protein expression profile in breast cancer patients
Yoon Jin Cha, Dooreh Kim, Soong June Bae, Sung Gwe Ahn, Joon Jeong, Min Kyung Cho, Pill Sun Paik, Tae-Kyung Yoo, Woo-Chan Park, Chang Ik Yoon, Elda Tagliabue
PLOS ONE.2021; 16(5): e0250986. CrossRef
- The future is now: advancing p53 immunohistochemistry in Barrett's oesophagus and its implication for the everyday pathologist
- Morule-like features in pulmonary adenocarcinoma associated with epidermal growth factor receptor mutations: two case reports with targeted next-generation sequencing analysis
- Yoo Jin Lee, Harim Oh, Eojin Kim, Bokyung Ahn, Jeong Hyeon Lee, Youngseok Lee, Yang Seok Chae, Chul Hwan Kim
- J Pathol Transl Med. 2020;54(1):119-122. Published online November 1, 2019
- DOI: https://doi.org/10.4132/jptm.2019.09.30
- 6,564 View
- 138 Download
- 2 Web of Science
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - Morules, or morule-like features, can be identified in benign and malignant lesions in various organs. Morular features are unusual in pulmonary adenocarcinoma cases with only 26 cases reported to date. Here, we describe two cases of pulmonary adenocarcinoma with morule-like features in Korean women. One patient had a non-mucinous-type adenocarcinoma in situ and the other had an acinarpredominant adenocarcinoma with a micropapillary component. Both patients showed multiple intra-alveolar, nodular, whorled proliferative foci composed of atypical spindle cells with eosinophilic cytoplasm. Targeted next-generation sequencing was performed on DNA extracted from formalin-fixed paraffin-embedded samples of the tumors. Results showed unusual epidermal growth factor receptor (EGFR) mutations, which are associated with drug resistance to EGFR tyrosine kinase inhibitors, revealing the importance of identifying morule-like features in pulmonary adenocarcinoma and the need for additional study, since there are few reported cases.
-
Citations
Citations to this article as recorded by- Pulmonary adenocarcinoma in situ with morule - like components: A surgical case report
Mitsuteru Yosida, Mitsuru Tomita, Naoya Kawakita, Teruki Shimizu, Ryou Yamada, Hiromitsu Takizawa, Hisanori Uehara
Respiratory Medicine Case Reports.2024; 48: 102008. CrossRef - Clinicopathological, Radiological, and Molecular Features of Primary Lung Adenocarcinoma with Morule-Like Components
Li-Li Wang, Li Ding, Peng Zhao, Jing-Jing Guan, Xiao-Bin Ji, Xiao-Li Zhou, Shi-Hong Shao, Yu-Wei Zou, Wei-Wei Fu, Dong-Liang Lin, Dong Pan
Disease Markers.2021; 2021: 1. CrossRef
- Pulmonary adenocarcinoma in situ with morule - like components: A surgical case report
- Utility of BRAF VE1 Immunohistochemistry as a Screening Tool for Colorectal Cancer Harboring BRAF V600E Mutation
- Jeong-Hwa Kwon, Byung-Kwan Jeong, Yong Sik Yoon, Chang Sik Yu, Jihun Kim
- J Pathol Transl Med. 2018;52(3):157-163. Published online March 29, 2018
- DOI: https://doi.org/10.4132/jptm.2018.03.28
- 9,195 View
- 206 Download
- 12 Web of Science
- 8 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
BRAF mutation has been recognized as an important biomarker of colorectal cancer (CRC) for targeted therapy and prognosis prediction. However, sequencing for every CRC case is not cost-effective. An antibody specific for BRAF V600E mutant protein has been introduced, and we thus examined the utility of BRAF VE1 immunohistochemistry for evaluating BRAF mutations in CRC.
Methods
Fifty-one BRAF-mutated CRCs and 100 age and sexmatched BRAF wild-type CRCs between 2005 and 2015 were selected from the archives of Asan Medical Center. Tissue microarrays were constructed and stained with BRAF VE1 antibody.
Results
Forty-nine of the 51 BRAF-mutant CRCs (96.1%) showed more than moderate cytoplasmic staining, except for two weakly stained cases. Six of 100 BRAF wild-type cases also stained positive with BRAF VE1 antibody; four stained weakly and two stained moderately. Normal colonic crypts showed nonspecific weak staining, and a few CRC cases exhibited moderate nuclear reactivity (3 BRAF-mutant and 10 BRAF wild-type cases). BRAF-mutated CRC patients had higher pathologic stages and worse survival than BRAF wild-type patients.
Conclusions
BRAF VE1 immunohistochemistry showed high sensitivity and specificity, but occasional nonspecific staining in tumor cell nuclei and normal colonic crypts may limit their routine clinical use. Thus, BRAF VE1 immunohistochemistry may be a useful screening tool for BRAF V600E mutation in CRCs, provided that additional sequencing studies can be done to confirm the mutation in BRAF VE1 antibody-positive cases. -
Citations
Citations to this article as recorded by- Next-Generation Sequencing in Oncology—A Guiding Compass for Targeted Therapy and Emerging Applications
Laurenția Nicoleta Galeș, Mihai-Andrei Păun, Ioana Butnariu, Laurentiu Simion, Loredana Sabina Cornelia Manolescu, Oana Gabriela Trifănescu, Rodica Maricela Anghel
International Journal of Molecular Sciences.2025; 26(7): 3123. CrossRef - A multi-omic analysis reveals a predictive value of tertiary lymphoid structures in improving the prognosis of colorectal cancer patients with BRAF mutation
Chao Qin, Shumin Cheng, Jingyun Ma, Lujing Li, Yun Leng, Lei Zheng, Huiying Chen, Hui Mo, Shi Li, Yuhong Liang, Yi Zhang, Wenxia Li, Jing Liang, Yuxuan Liu, Junxuan Mai, Linlin Hou, Di Wang, Ke Zhu, Bihui Huang
Frontiers in Immunology.2025;[Epub] CrossRef - The evolving landscape of tissue‐agnostic therapies in precision oncology
Vivek Subbiah, Mohamed A. Gouda, Bettina Ryll, Howard A. Burris, Razelle Kurzrock
CA: A Cancer Journal for Clinicians.2024; 74(5): 433. CrossRef - Deciphering the Role of BRAFV600E Immunohistochemistry in Breast Lesions: A Comprehensive Review
Simran Khan, Arvind Bhake, Shakti Sagar
Cureus.2024;[Epub] CrossRef - Immunohistochemistry as a Surrogate Marker of Underlying Molecular Derangements in Sporadic Colorectal Carcinoma in Children – A Series of Three Cases
Priyanka Maity, Aniket Halder, Ranajoy Ghosh, Uttara Chatterjee, Shibsankar Barman, Ruchirendra Sarkar
Fetal and Pediatric Pathology.2022; 41(1): 98. CrossRef - Risk assessment and genetic counseling for Lynch syndrome – Practice resource of the National Society of Genetic Counselors and the Collaborative Group of the Americas on Inherited Gastrointestinal Cancer
Spring Holter, Michael J. Hall, Heather Hampel, Kory Jasperson, Sonia S. Kupfer, Joy Larsen Haidle, Maureen E. Mork, Selvi Palaniapppan, Leigha Senter, Elena M. Stoffel, Scott M. Weissman, Matthew B. Yurgelun
Journal of Genetic Counseling.2022; 31(3): 568. CrossRef - Current concepts in ameloblastoma-targeted therapies in B-raf proto-oncogene serine/threonine kinase V600E mutation: Systematic review
Rogelio González-González, Sandra López-Verdín, Jesús Lavalle-Carrasco, Nelly Molina-Frechero, Mario Isiordia-Espinoza, Ramón G Carreón-Burciaga, Ronell Bologna-Molina
World Journal of Clinical Oncology.2020; 11(1): 31. CrossRef - Genetic and histopathological analysis of a case of primary intraosseous carcinoma, NOS with features of both ameloblastic carcinoma and squamous cell carcinoma
Akane Yukimori, Maiko Tsuchiya, Akane Wada, Yasuyuki Michi, Kou Kayamori, Kei Sakamoto, Tohru Ikeda
World Journal of Surgical Oncology.2020;[Epub] CrossRef
- Next-Generation Sequencing in Oncology—A Guiding Compass for Targeted Therapy and Emerging Applications
- Molecular Screening of Small Biopsy Samples Using Next-Generation Sequencing in Korean Patients with Advanced Non-small Cell Lung Cancer: Korean Lung Cancer Consortium (KLCC-13-01)
- Bo Mi Ku, Mi Hwa Heo, Joo-Hang Kim, Byoung Chul Cho, Eun Kyung Cho, Young Joo Min, Ki Hyeong Lee, Jong-Mu Sun, Se-Hoon Lee, Jin Seok Ahn, Keunchil Park, Tae Jung Kim, Ho Yun Lee, Hojoong Kim, Kyung-Jong Lee, Myung-Ju Ahn
- J Pathol Transl Med. 2018;52(3):148-156. Published online March 26, 2018
- DOI: https://doi.org/10.4132/jptm.2018.03.12
- 10,689 View
- 315 Download
- 16 Web of Science
- 14 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
Non-small cell lung cancer (NSCLC) is a common type of cancer with poor prognosis. As individual cancers exhibit unique mutation patterns, identifying and characterizing gene mutations in NSCLC might help predict patient outcomes and guide treatment. The aim of this study was to evaluate the clinical adequacy of molecular testing using next-generation sequencing (NGS) for small biopsy samples and characterize the mutational landscape of Korean patients with advanced NSCLC.
Methods
DNA was extracted from small biopsy samples of 162 patients with advanced NSCLC. Targeted NGS of genomic alterations was conducted using Ion AmpliSeq Cancer Hotspot Panel v2.
Results
The median age of patients was 64 years (range, 32 to 83 years) and the majority had stage IV NSCLC at the time of cancer diagnosis (90%). Among the 162 patients, 161 patients (99.4%) had novel or hotspot mutations (range, 1 to 21 mutated genes). Mutations were found in 41 genes. Three of the most frequently mutated genes were TP53 (151, 93.2%), KDR (104, 64.2%), and epidermal growth factor receptor (EGFR; 69, 42.6%). We also observed coexistence of EGFR and other oncogene (such as KRAS, PIC3CA, PTEN, and STK11) mutations. Given that 69.6% (48/69) of EGFR mutant patients were treated with EGFR tyrosine kinase inhibitors, EGFR mutant status had higher prognostic ability in this study.
Conclusions
These results suggest that targeted NGS using small biopsy samples is feasible and allows for the detection of both common and rare mutations in NSCLC. -
Citations
Citations to this article as recorded by- The clinical relevance of surgical specimens for RNA sequencing in lung cancer: a cohort study
Jung Seop Eom, Soo Han Kim, Kyungbin Kim, Ahrong Kim, Hyo Yeong Ahn, Jeongha Mok, Jeong Su Cho, Min Ki Lee, Ju Sun Song, Mi-Hyun Kim
Frontiers in Oncology.2024;[Epub] CrossRef - PTEN, PTENP1, microRNAs, and ceRNA Networks: Precision Targeting in Cancer Therapeutics
Glena Travis, Eileen M. McGowan, Ann M. Simpson, Deborah J. Marsh, Najah T. Nassif
Cancers.2023; 15(20): 4954. CrossRef - Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis
Barbara Melosky, Kato Kambartel, Maik Häntschel, Margherita Bennetts, Dana J. Nickens, Julia Brinkmann, Antonin Kayser, Michael Moran, Federico Cappuzzo
Molecular Diagnosis & Therapy.2022; 26(1): 7. CrossRef - Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2022; 56(5): 249. CrossRef - Suitability of transbronchial brushing cytology specimens for next‐generation sequencing in peripheral lung cancer
Naoki Furuya, Shingo Matsumoto, Kazutaka Kakinuma, Kei Morikawa, Takeo Inoue, Hisashi Saji, Koichi Goto, Masamichi Mineshita
Cancer Science.2021; 112(1): 380. CrossRef - KLHL38 involvement in non-small cell lung cancer progression via activation of the Akt signaling pathway
Yitong Xu, Chenglong Wang, Xizi Jiang, Yao Zhang, Hongbo Su, Jun Jiang, Hongjiu Ren, Xueshan Qiu
Cell Death & Disease.2021;[Epub] CrossRef - Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
Journal of Pathology and Translational Medicine.2021; 55(3): 181. CrossRef - Targeting non-small cell lung cancer: driver mutation beyond epidermal growth factor mutation and anaplastic lymphoma kinase fusion
Quincy S. Chu
Therapeutic Advances in Medical Oncology.2020;[Epub] CrossRef Concomitant Mutations in EGFR 19Del/L858R Mutation and Their Association with Response to EGFR-TKIs in NSCLC Patients
Hengrui Liang, Caichen Li, Yi Zhao, Shen Zhao, Jun Huang, Xiuyu Cai, Bo Cheng, Shan Xiong, Jianfu Li, Wei Wang, Changbin Zhu, Weiwei Li, Jianxing He, Wenhua Liang
Cancer Management and Research.2020; Volume 12: 8653. CrossRef- Prognostic role of Rab27A and Rab27B expression in patients with non‐small cell lung carcinoma
Hyun Min Koh, Dae Hyun Song
Thoracic Cancer.2019; 10(2): 143. CrossRef - PD‐L1 expression in ROS1‐rearranged non‐small cell lung cancer: A study using simultaneous genotypic screening of EGFR, ALK, and ROS1
Jongmin Lee, Chan Kwon Park, Hyoung‐Kyu Yoon, Young Jo Sa, In Sook Woo, Hyo Rim Kim, Sue Youn Kim, Tae‐Jung Kim
Thoracic Cancer.2019; 10(1): 103. CrossRef - Targeted Next-Generation Sequencing Validates the Use of Diagnostic Biopsies as a Suitable Alternative to Resection Material for Mutation Screening in Colorectal Cancer
Hersh A. Ham-Karim, Henry Okuchukwu Ebili, Kirsty Manger, Wakkas Fadhil, Narmeen S. Ahmad, Susan D. Richman, Mohammad Ilyas
Molecular Diagnosis & Therapy.2019; 23(3): 383. CrossRef - Small lung tumor biopsy samples are feasible for high quality targeted next generation sequencing
Hidenori Kage, Shinji Kohsaka, Aya Shinozaki‐Ushiku, Yoshihisa Hiraishi, Jiro Sato, Kazuhiro Nagayama, Tetsuo Ushiku, Daiya Takai, Jun Nakajima, Kiyoshi Miyagawa, Hiroyuki Aburatani, Hiroyuki Mano, Takahide Nagase
Cancer Science.2019; 110(8): 2652. CrossRef - PTEN in Lung Cancer: Dealing with the Problem, Building on New Knowledge and Turning the Game Around
Anastasios Gkountakos, Giulia Sartori, Italia Falcone, Geny Piro, Ludovica Ciuffreda, Carmine Carbone, Giampaolo Tortora, Aldo Scarpa, Emilio Bria, Michele Milella, Rafael Rosell, Vincenzo Corbo, Sara Pilotto
Cancers.2019; 11(8): 1141. CrossRef
- The clinical relevance of surgical specimens for RNA sequencing in lung cancer: a cohort study
- Molecular Testing of Brain Tumor
- Sung-Hye Park, Jaekyung Won, Seong-Ik Kim, Yujin Lee, Chul-Kee Park, Seung-Ki Kim, Seung-Hong Choi
- J Pathol Transl Med. 2017;51(3):205-223. Published online May 12, 2017
- DOI: https://doi.org/10.4132/jptm.2017.03.08
- 35,577 View
- 1,175 Download
- 40 Web of Science
- 45 Crossref
-
 Abstract
Abstract
 PDF
PDF - The World Health Organization (WHO) classification of central nervous system (CNS) tumors was revised in 2016 with a basis on the integrated diagnosis of molecular genetics. We herein provide the guidelines for using molecular genetic tests in routine pathological practice for an accurate diagnosis and appropriate management. While astrocytomas and IDH-mutant (secondary) glioblastomas are characterized by the mutational status of IDH, TP53, and ATRX, oligodendrogliomas have a 1p/19q codeletion and mutations in IDH, CIC, FUBP1, and the promoter region of telomerase reverse transcriptase (TERTp). IDH-wildtype (primary) glioblastomas typically lack mutations in IDH, but are characterized by copy number variations of EGFR, PTEN, CDKN2A/B, PDGFRA, and NF1 as well as mutations of TERTp. High-grade pediatric gliomas differ from those of adult gliomas, consisting of mutations in H3F3A, ATRX, and DAXX, but not in IDH genes. In contrast, well-circumscribed low-grade neuroepithelial tumors in children, such as pilocytic astrocytoma, pleomorphic xanthoastrocytoma, and ganglioglioma, often have mutations or activating rearrangements in the BRAF, FGFR1, and MYB genes. Other CNS tumors, such as ependymomas, neuronal and glioneuronal tumors, embryonal tumors, meningothelial, and other mesenchymal tumors have important genetic alterations, many of which are diagnostic, prognostic, and predictive markers and therapeutic targets. Therefore, the neuropathological evaluation of brain tumors is increasingly dependent on molecular genetic tests for proper classification, prediction of biological behavior and patient management. Identifying these gene abnormalities requires cost-effective and high-throughput testing, such as next-generation sequencing. Overall, this paper reviews the global guidelines and diagnostic algorithms for molecular genetic testing of brain tumors.
-
Citations
Citations to this article as recorded by- Navigating rare vascular brain tumors: A retrospective observational study
Sana Ahuja, Dipanker S Mankotia, Naveen Kumar, Vyomika Teckchandani, Sufian Zaheer
Cancer Research, Statistics, and Treatment.2025; 8(2): 92. CrossRef - Author’s reply to Dagar and Mallick
Sana Ahuja, Sufian Zaheer
Cancer Research, Statistics, and Treatment.2025; 8(3): 245. CrossRef - The Clinical Utility of Cell-Free DNA in Brain Tumor Management: A Comprehensive Review
Qama Abuhassan, Hanan Hassan Ahmed, Radhwan Abdul Kareem, Soumya V. Menon, Priya Priyadarshini Nayak, J. Bethanney Janney, Vimal Arora, Aashna Sinha, Saif Aldeen Jaber, Hayder Naji Sameer, Ahmed Yaseen, Zainab H. Athab, Mohaned Adil
Journal of Molecular Neuroscience.2025;[Epub] CrossRef - PTEN regulates expression of its pseudogene in glioblastoma cells in DNA methylation-dependent manner
Tatyana F. Kovalenko, Bhupender Yadav, Ksenia S. Anufrieva, Tatyana D. Larionova, Tatiana E. Aksinina, Yaroslav A. Latyshev, Soniya Bastola, Michail I. Shakhparonov, Amit Kumar Pandey, Marat S. Pavlyukov
Biochimie.2024; 219: 74. CrossRef - CDKN2A/B deletion in IDH-mutant astrocytomas: An evaluation by Fluorescence in-situ hybridization
Manali Ranade, Sridhar Epari, Omshree Shetty, Sandeep Dhanavade, Sheetal Chavan, Ayushi Sahay, Arpita Sahu, Prakash Shetty, Aliasgar Moiyadi, Vikash Singh, Archya Dasgupta, Abhishek Chatterjee, Sadhana Kannan, Tejpal Gupta
Journal of Neuro-Oncology.2024; 167(1): 189. CrossRef - Central neurocytoma exhibits radial glial cell signatures with FGFR3 hypomethylation and overexpression
Yeajina Lee, Tamrin Chowdhury, Sojin Kim, Hyeon Jong Yu, Kyung-Min Kim, Ho Kang, Min-Sung Kim, Jin Wook Kim, Yong-Hwy Kim, So Young Ji, Kihwan Hwang, Jung Ho Han, Jinha Hwang, Seong-Keun Yoo, Kyu Sang Lee, Gheeyoung Choe, Jae-Kyung Won, Sung-Hye Park, Yon
Experimental & Molecular Medicine.2024; 56(4): 975. CrossRef - Liquid biopsy in brain tumors: Potential for impactful clinical applications
Tania Eid, Lina Ghandour, Joseph Abi Ghanem, Hazem Assi, Rami Mahfouz
Human Gene.2024; 42: 201333. CrossRef - Diagnostic challenges in complicated case of glioblastoma
Tatiana Aghova, Halka Lhotska, Libuse Lizcova, Karla Svobodova, Lucie Hodanova, Karolina Janeckova, Kim Vucinic, Martin Gregor, Dora Konecna, Filip Kramar, Jiri Soukup, David Netuka, Zuzana Zemanova
Pathology and Oncology Research.2024;[Epub] CrossRef - Drugging Hijacked Kinase Pathways in Pediatric Oncology: Opportunities and Current Scenario
Marina Ferreira Candido, Mariana Medeiros, Luciana Chain Veronez, David Bastos, Karla Laissa Oliveira, Julia Alejandra Pezuk, Elvis Terci Valera, María Sol Brassesco
Pharmaceutics.2023; 15(2): 664. CrossRef - BrainBase: a curated knowledgebase for brain diseases
Lin Liu, Yang Zhang, Guangyi Niu, Qianpeng Li, Zhao Li, Tongtong Zhu, Changrui Feng, Xiaonan Liu, Yuansheng Zhang, Tianyi Xu, Ruru Chen, Xufei Teng, Rongqin Zhang, Dong Zou, Lina Ma, Zhang Zhang
Nucleic Acids Research.2022; 50(D1): D1131. CrossRef - A heat shock protein 90 inhibitor reduces oncoprotein expression and induces cell death in heterogeneous glioblastoma cells with EGFR, PDGFRA, CDK4, and NF1 aberrations
Kuan-Ta Ho, Pei-Fan Chen, Jian-Ying Chuang, Po-Wu Gean, Yuan-Shuo Hsueh
Life Sciences.2022; 288: 120176. CrossRef - Bicentric validation of the navigated transcranial magnetic stimulation motor risk stratification model
Tizian Rosenstock, Levin Häni, Ulrike Grittner, Nicolas Schlinkmann, Meltem Ivren, Heike Schneider, Andreas Raabe, Peter Vajkoczy, Kathleen Seidel, Thomas Picht
Journal of Neurosurgery.2022; 136(4): 1194. CrossRef - Foramen magnum meningioma presented as cervical myelopathy in a pregnant COVID-19 patient: A case report
Olivia Josephine Wijaya, Djohan Ardiansyah
Annals of Medicine and Surgery.2022; 77: 103647. CrossRef - The telomere maintenance mechanism spectrum and its dynamics in gliomas
Sojin Kim, Tamrin Chowdhury, Hyeon Jong Yu, Jee Ye Kahng, Chae Eun Lee, Seung Ah. Choi, Kyung-Min Kim, Ho Kang, Joo Ho Lee, Soon-Tae Lee, Jae-Kyung Won, Kyung Hyun Kim, Min-Sung Kim, Ji Yeoun Lee, Jin Wook Kim, Yong-Hwy Kim, Tae Min Kim, Seung Hong Choi,
Genome Medicine.2022;[Epub] CrossRef - A review of predictive, prognostic and diagnostic biomarkers for brain tumours: towards personalised and targeted cancer therapy
Ernest Osei, Pascale Walters, Olivia Masella, Quinton Tennant, Amber Fishwick, Eugenia Dadzie, Anmol Bhangu, Johnson Darko
Journal of Radiotherapy in Practice.2021; 20(1): 83. CrossRef - Use of a novel navigable tubular retractor system in 1826 minimally invasive parafascicular surgery (MIPS) cases involving deep-seated brain tumors, hemorrhages and malformations
Martina M. Cartwright, Penny Sekerak, Joseph Mark, Julian Bailes
Interdisciplinary Neurosurgery.2021; 23: 100919. CrossRef - Disentangling the therapeutic tactics in GBM: From bench to bedside and beyond
S. Daisy Precilla, Shreyas S. Kuduvalli, Anitha Thirugnanasambandhar Sivasubramanian
Cell Biology International.2021; 45(1): 18. CrossRef - Sequencing of a central nervous system tumor demonstrates cancer transmission in an organ transplant
Marie-Claude Gingras, Aniko Sabo, Maria Cardenas, Abbas Rana, Sadhna Dhingra, Qingchang Meng, Jianhong Hu, Donna M Muzny, Harshavardhan Doddapaneni, Lesette Perez, Viktoriya Korchina, Caitlin Nessner, Xiuping Liu, Hsu Chao, John Goss, Richard A Gibbs
Life Science Alliance.2021; 4(9): e202000941. CrossRef - Radiogenomics of Gliomas
Chaitra Badve, Sangam Kanekar
Radiologic Clinics of North America.2021; 59(3): 441. CrossRef - Chemical analysis of the human brain by imaging mass spectrometry
Akhila Ajith, Yeswanth Sthanikam, Shibdas Banerjee
The Analyst.2021; 146(18): 5451. CrossRef - Correlation between Stage and Histopathological Features and Clinical Outcomes in Patients with Glioma Tumors
Andre Lona, Alfansuri Kadri, Irina Kemala Nasution
Open Access Macedonian Journal of Medical Sciences.2021; 9(T3): 262. CrossRef - Pediatric Glioma: An Update of Diagnosis, Biology, and Treatment
Yusuke Funakoshi, Nobuhiro Hata, Daisuke Kuga, Ryusuke Hatae, Yuhei Sangatsuda, Yutaka Fujioka, Kosuke Takigawa, Masahiro Mizoguchi
Cancers.2021; 13(4): 758. CrossRef - Discovery of clinically relevant fusions in pediatric cancer
Stephanie LaHaye, James R. Fitch, Kyle J. Voytovich, Adam C. Herman, Benjamin J. Kelly, Grant E. Lammi, Jeremy A. Arbesfeld, Saranga Wijeratne, Samuel J. Franklin, Kathleen M. Schieffer, Natalie Bir, Sean D. McGrath, Anthony R. Miller, Amy Wetzel, Katheri
BMC Genomics.2021;[Epub] CrossRef - Clinicopathological correlation of glioma patients with respect to immunohistochemistry markers: A prospective study of 115 patients in a Tertiary Care Hospital in North India
Gitanshu Dahuja, Ashok Gupta, Arpita Jindal, Gaurav Jain, Santosh Sharma, Arvind Kumar
Asian Journal of Neurosurgery.2021; 16(04): 732. CrossRef - Role of Extracellular Vesicles in Glioma Progression: Deciphering Cellular Biological Processes to Clinical Applications
Rashmi Rana, Shikha Joon, Kirti Chauhan, Vaishnavi Rathi, Nirmal Kumar Ganguly, Chandni Kumari, Dharmendra Kumar Yadav
Current Topics in Medicinal Chemistry.2021; 21(8): 696. CrossRef - A comprehensive overview on the molecular biology of human glioma: what the clinician needs to know
P. D. Delgado-López, P. Saiz-López, R. Gargini, E. Sola-Vendrell, S. Tejada
Clinical and Translational Oncology.2020; 22(11): 1909. CrossRef - Use of Fluorescence In Situ Hybridization (FISH) in Diagnosis and Tailored Therapies in Solid Tumors
Natalia Magdalena Chrzanowska, Janusz Kowalewski, Marzena Anna Lewandowska
Molecules.2020; 25(8): 1864. CrossRef - From Banding to BAM Files
Adrian M. Dubuc
Surgical Pathology Clinics.2020; 13(2): 343. CrossRef - Mutation profiling of anaplastic ependymoma grade III by Ion Proton next generation DNA sequencing
Ejaz Butt, Sabra Alyami, Tahani Nageeti, Muhammad Saeed, Khalid AlQuthami, Abdellatif Bouazzaoui, Mohammad Athar, Zainularifeen Abduljaleel, Faisal Al-Allaf, Mohiuddin Taher
F1000Research.2020; 8: 613. CrossRef - Assessment of the non-linear optical behavior of cells for discrimination between normal and malignant glial cells
Soraya Emamgholizadeh Minaei, Alireza Ghader, Ali Abbasian Ardakani, Samideh Khoei, Mohammad Hosein Majles Ara
Laser Physics.2020; 30(12): 125601. CrossRef - Mutation profiling of anaplastic ependymoma grade III by Ion Proton next generation DNA sequencing
Muhammad Butt, Sabra Alyami, Tahani Nageeti, Muhammad Saeed, Khalid AlQuthami, Abdellatif Bouazzaoui, Mohammad Athar, Zainularifeen Abduljaleel, Faisal Al-Allaf, Mohiuddin Taher
F1000Research.2019; 8: 613. CrossRef - A Multiplex Quantitative Reverse Transcription Polymerase Chain Reaction Assay for the Detection of KIAA1549–BRAF Fusion Transcripts in Formalin-Fixed Paraffin-Embedded Pilocytic Astrocytomas
David Bret, Valentin Chappuis, Delphine Poncet, François Ducray, Karen Silva, Fabrice Mion, Alexandre Vasiljevic, Carole Ferraro-Peyret, Carmine Mottolese, Pierre Leblond, Mathieu Gabut, Didier Frappaz, Nathalie Streichenberger, David Meyronet, Pierre-Pau
Molecular Diagnosis & Therapy.2019; 23(4): 537. CrossRef - A PTPmu Biomarker is Associated with Increased Survival in Gliomas
Mette L. Johansen, Jason Vincent, Haley Gittleman, Sonya E. L. Craig, Marta Couce, Andrew E. Sloan, Jill S. Barnholtz-Sloan, Susann M. Brady-Kalnay
International Journal of Molecular Sciences.2019; 20(10): 2372. CrossRef -
ATRX Mutations in Pineal Parenchymal Tumors of Intermediate Differentiation
Haydee Martínez, Michelle Nagurney, Zi-Xuan Wang, Charles G Eberhart, Christopher M Heaphy, Mark T Curtis, Fausto J Rodriguez
Journal of Neuropathology & Experimental Neurology.2019; 78(8): 703. CrossRef - The role of fibroblast growth factors and their receptors in gliomas: the mutations involved
Vasiliki Georgiou, Vasiliki Gkretsi
Reviews in the Neurosciences.2019; 30(5): 543. CrossRef - The Role of Next Generation Sequencing in Diagnosis of Brain Tumors: A Review Study
Sadegh Shirian, Yahya Daneshbod, Saranaz Jangjoo, Amir Ghaemi, Arash Goodarzi, Maryam Ghavideldarestani, Ahmad Emadi, Arman Ai, Akbar Ahmadi, Jafar Ai
Archives of Neuroscience.2019;[Epub] CrossRef - Applied Precision Cancer Medicine in Neuro-Oncology
H. Taghizadeh, L. Müllauer, J. Furtner, J. A. Hainfellner, C. Marosi, M. Preusser, G. W. Prager
Scientific Reports.2019;[Epub] CrossRef - Comparative genomic analysis of driver mutations in matched primary and recurrent meningiomas
Joshua Loewenstern, John Rutland, Corey Gill, Hanane Arib, Margaret Pain, Melissa Umphlett, Yayoi Kinoshita, Russell McBride, Michael Donovan, Robert Sebra, Joshua Bederson, Mary Fowkes, Raj Shrivastava
Oncotarget.2019; 10(37): 3506. CrossRef - Next generation DNA sequencing of atypical choroid plexus papilloma of brain: Identification of novel mutations in a female patient by Ion Proton
Mohiuddin Taher, Amal Hassan, Muhammad Saeed, Raid Jastania, Tahani Nageeti, Hisham Alkhalidi, Ghida Dairi, Zainularifeen Abduljaleel, Mohammad Athar, Abdellatif Bouazzaoui, Wafa El‑Bjeirami, Faisal Al‑Allaf
Oncology Letters.2019;[Epub] CrossRef - Pure mechanistic analysis of additive neuroprotective effects between baicalin and jasminoidin in ischemic stroke mice
Peng-qian Wang, Qiong Liu, Wen-juan Xu, Ya-nan Yu, Ying-ying Zhang, Bing Li, Jun Liu, Zhong Wang
Acta Pharmacologica Sinica.2018; 39(6): 961. CrossRef - The Smad4/PTEN Expression Pattern Predicts Clinical Outcomes in Colorectal Adenocarcinoma
Yumin Chung, Young Chan Wi, Yeseul Kim, Seong Sik Bang, Jung-Ho Yang, Kiseok Jang, Kyueng-Whan Min, Seung Sam Paik
Journal of Pathology and Translational Medicine.2018; 52(1): 37. CrossRef - Primary brain tumours in adults
Sarah Lapointe, Arie Perry, Nicholas A Butowski
The Lancet.2018; 392(10145): 432. CrossRef - Epidemiology and Overview of Gliomas
Mary Elizabeth Davis
Seminars in Oncology Nursing.2018; 34(5): 420. CrossRef - Toward Precision Medicine: Promising Areas of Research in Glioma
Mary Elizabeth Davis, Dana Bossert, Malbora Manne
Seminars in Oncology Nursing.2018; 34(5): 569. CrossRef - Molecular Basis of Pediatric Brain Tumors
Alexia Klonou, Christina Piperi, Antonios N. Gargalionis, Athanasios G. Papavassiliou
NeuroMolecular Medicine.2017; 19(2-3): 256. CrossRef
- Navigating rare vascular brain tumors: A retrospective observational study
- Molecular Testing of Lymphoproliferative Disorders: Current Status and Perspectives
- Yoon Kyung Jeon, Sun Och Yoon, Jin Ho Paik, Young A Kim, Bong Kyung Shin, Hyun-Jung Kim, Hee Jeong Cha, Ji Eun Kim, Jooryung Huh, Young-Hyeh Ko
- J Pathol Transl Med. 2017;51(3):224-241. Published online May 10, 2017
- DOI: https://doi.org/10.4132/jptm.2017.04.09
- 21,818 View
- 710 Download
- 13 Web of Science
- 16 Crossref
-
 Abstract
Abstract
 PDF
PDF - Molecular pathologic testing plays an important role for the diagnosis, prognostication and decision of treatment strategy in lymphoproliferative disease. Here, we briefly review the molecular tests currently used for lymphoproliferative disease and those which will be implicated in clinical practice in the near future. Specifically, this guideline addresses the clonality test for B- and T-cell proliferative lesions, molecular cytogenetic tests for malignant lymphoma, determination of cell-of-origin in diffuse large B-cell lymphoma, and molecular genetic alterations incorporated in the 2016 revision of the World Health Organization classification of lymphoid neoplasms. Finally, a new perspective on the next-generation sequencing for diagnostic, prognostic, and therapeutic purpose in malignant lymphoma will be summarized.
-
Citations
Citations to this article as recorded by- Pediatric lymphoproliferative disorders – Emerging insights and management: A narrative review
Emmanuel Ifeanyi Obeagu
Medicine.2026; 105(4): e47367. CrossRef - Presence of minimal residual disease determined by next-generation sequencing is not a reliable prognostic biomarker in children with acute lymphoblastic leukemia
Elizabeta Krstevska Bozhinovikj, Nadica Matevska-Geshkovska, Marija Staninova Stojovska, Emilija Gjorgievska, Aleksandra Jovanovska, Nevenka Ridova, Irina Panovska Stavridis, Svetlana Kocheva, Aleksandar Dimovski
Leukemia & Lymphoma.2025; 66(6): 1121. CrossRef - Haematogenous seeding in mycosis fungoides and Sézary syndrome: current evidence and clinical implications
Robert Gniadecki, Emmanuella Guenova, Christiane Querfeld, Jan P Nicolay, Julia Scarisbrick, Lubomir Sokol
British Journal of Dermatology.2025; 192(3): 381. CrossRef - Exploring External Quality Control Methods for PCR–Polyacrylamide Gel Electrophoresis–Based Lymphocyte Receptor Gene Rearrangement Assays in Korea
Jieun Kim, Ho Hyun Song, Soobin Chae, GeonWoo Choi, Jeong Won Shin
Journal of Laboratory Medicine and Quality Assurance.2025; 47(2): 43. CrossRef - Laboratory analysis of 182 cases of B-cell lymphoproliferative disorders other than typical chronic lymphocytic leukemia: Single-center study
Shams Salah Mahdi, Nuha Abd Ali Al-Sarai
Iraqi Journal of Hematology.2025; 14(2): 218. CrossRef - Assessment of Bone Marrow Involvement in B‐Cell non‐Hodgkin Lymphoma Using Immunoglobulin Gene Rearrangement Analysis with Next‐Generation Sequencing
Min Ji Jeon, Eun Sang Yu, Dae Sik Kim, Chul Won Choi, Ha Nui Kim, Jung Ah Kwon, Soo‐Young Yoon, Jung Yoon
Journal of Clinical Laboratory Analysis.2024;[Epub] CrossRef - Thymus and lung mucosa-associated lymphoid tissue lymphoma with adenocarcinoma of the lung: a case report and literature review
Yu Pang, Daosheng Li, Yiqian Chen, Qinqin Liu, Yuheng Wu, Qingliang Teng, Yuyu Liu
World Journal of Surgical Oncology.2023;[Epub] CrossRef - Development and implementation of an automated and highly accurate reporting process for NGS-based clonality testing
Sean T. Glenn, Phillip M. Galbo, Jesse D. Luce, Kiersten Marie Miles, Prashant K. Singh, Manuel J. Glynias, Carl Morrison
Oncotarget.2023; 14(1): 450. CrossRef - A comparison of capillary electrophoresis and next-generation sequencing in the detection of immunoglobulin heavy chain H and light chain κ gene rearrangements in the diagnosis of classic hodgkin’s lymphoma
Juan-Juan Zhang, Yu-Xin Xie, Li-Lin Luo, Xuan-Tao Yang, Yi-Xing Wang, Yue Cao, Zheng-Bo Long, Wan-Pu Wang
Bioengineered.2022; 13(3): 5868. CrossRef - Lymphoproliferative disorder involving body fluid: diagnostic approaches and roles of ancillary studies
Jiwon Koh, Sun Ah Shin, Ji Ae Lee, Yoon Kyung Jeon
Journal of Pathology and Translational Medicine.2022; 56(4): 173. CrossRef - Diagnostic Workup of Primary Cutaneous B Cell Lymphomas: A Clinician's Approach
Giulia Tadiotto Cicogna, Martina Ferranti, Mauro Alaibac
Frontiers in Oncology.2020;[Epub] CrossRef - Kappa and lambda immunohistochemistry and in situ hybridization in the evaluation of atypical cutaneous lymphoid infiltrates
Alexandra C. Hristov, Nneka I. Comfere, Claudia I. Vidal, Uma Sundram
Journal of Cutaneous Pathology.2020; 47(11): 1103. CrossRef - Primary lung mucosa-associated lymphoid tissue lymphoma accompanied by multiple sclerosis
Ke-Ke Yu, Lei Zhu, Ji-Kai Zhao, Rui-Ying Zhao, Yu-Chen Han
Chinese Medical Journal.2019; 132(13): 1625. CrossRef - Diagnostic accuracy of SOX11 immunohistochemistry in mantle cell lymphoma: A meta-analysis
Woojoo Lee, Eun Shin, Bo-Hyung Kim, Hyunchul Kim, Riccardo Dolcetti
PLOS ONE.2019; 14(11): e0225096. CrossRef - Views of dermatopathologists about clonality assays in the diagnosis of cutaneous T‐cell and B‐cell lymphoproliferative disorders
Nneka Comfere, Uma Sundram, Maria Yadira Hurley, Brian Swick
Journal of Cutaneous Pathology.2018; 45(1): 39. CrossRef - A Next-Generation Sequencing Primer—How Does It Work and What Can It Do?
Yuriy O. Alekseyev, Roghayeh Fazeli, Shi Yang, Raveen Basran, Thomas Maher, Nancy S. Miller, Daniel Remick
Academic Pathology.2018; 5: 2374289518766521. CrossRef
- Pediatric lymphoproliferative disorders – Emerging insights and management: A narrative review
- Good Laboratory Standards for Clinical Next-Generation Sequencing Cancer Panel Tests
- Jihun Kim, Woong-Yang Park, Nayoung K. D. Kim, Se Jin Jang, Sung-Min Chun, Chang-Ohk Sung, Jene Choi, Young-Hyeh Ko, Yoon-La Choi, Hyo Sup Shim, Jae-Kyung Won
- J Pathol Transl Med. 2017;51(3):191-204. Published online May 10, 2017
- DOI: https://doi.org/10.4132/jptm.2017.03.14
- 29,141 View
- 1,118 Download
- 36 Web of Science
- 37 Crossref
-
 Abstract
Abstract
 PDF
PDF - Next-generation sequencing (NGS) has recently emerged as an essential component of personalized cancer medicine due to its high throughput and low per-base cost. However, no sufficient guidelines for implementing NGS as a clinical molecular pathology test are established in Korea. To ensure clinical grade quality without inhibiting adoption of NGS, a taskforce team assembled by the Korean Society of Pathologists developed laboratory guidelines for NGS cancer panel testing procedures and requirements for clinical implementation of NGS. This consensus standard proposal consists of two parts: laboratory guidelines and requirements for clinical NGS laboratories. The laboratory guidelines part addressed several important issues across multistep NGS cancer panel tests including choice of gene panel and platform, sample handling, nucleic acid management, sample identity tracking, library preparation, sequencing, analysis and reporting. Requirements for clinical NGS tests were summarized in terms of documentation, validation, quality management, and other required written policies. Together with appropriate pathologist training and international laboratory standards, these laboratory standards would help molecular pathology laboratories to successfully implement NGS cancer panel tests in clinic. In this way, the oncology community would be able to help patients to benefit more from personalized cancer medicine.
-
Citations
Citations to this article as recorded by- Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis
Lydia A Schoenpflug, Aikaterini Chatzipli, Korsuk Sirinukunwattana, Susan Richman, Andrew Blake, James Robineau, Kirsten D Mertz, Clare Verrill, Simon J Leedham, Claire Hardy, Celina Whalley, Keara Redmond, Philip Dunne, Steven Walker, Andrew D Beggs, Ult
The Journal of Pathology.2025; 265(2): 184. CrossRef - Clinical Validation of Local Versus Commercial Genomic Testing in Cancer: A Comparison of Tissue and Plasma Concordance
Lucy G. Faulkner, Lynne Howells, Susann Lehman, Caroline Cowley, Zahirah Sidat, Jacqui Shaw, Anne L. Thomas
Cancer Investigation.2025; 43(2): 119. CrossRef - Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement
Bernhard Strasser, Sebastian Mustafa, Josef Seier, Erich Wimmer, Josef Tomasits
Diagnostics.2025; 15(6): 727. CrossRef - Pragmatic nationwide master observational trial based on genomic alterations in advanced solid tumors: KOrean Precision Medicine Networking Group Study of MOlecular profiling guided therapy based on genomic alterations in advanced Solid tumors (KOSMOS)-II
Sun Young Kim, Jee Hyun Kim, Tae-Yong Kim, Sook Ryun Park, Shinkyo Yoon, Soohyeon Lee, Se-Hoon Lee, Tae Min Kim, Sae-Won Han, Hye Ryun Kim, Hongseok Yun, Sejoon Lee, Jihun Kim, Yoon-La Choi, Kui Son Choi, Heejung Chae, Hyewon Ryu, Gyeong-Won Lee, Dae Youn
BMC Cancer.2024;[Epub] CrossRef - Reporting of somatic variants in clinical cancer care: recommendations of the Swiss Society of Molecular Pathology
Yann Christinat, Baptiste Hamelin, Ilaria Alborelli, Paolo Angelino, Valérie Barbié, Bettina Bisig, Heather Dawson, Milo Frattini, Tobias Grob, Wolfram Jochum, Ronny Nienhold, Thomas McKee, Matthias Matter, Edoardo Missiaglia, Francesca Molinari, Sacha Ro
Virchows Archiv.2024; 485(6): 1033. CrossRef - Acute myeloid leukemia and myelodysplastic neoplasms: clinical implications of myelodysplasia-related genes mutations and TP53 aberrations
Hyunwoo Kim, Ja Young Lee, Shinae Yu, Eunkyoung Yoo, Hye Ran Kim, Sang Min Lee, Won Sik Lee
Blood Research.2024;[Epub] CrossRef - Validation and Clinical Application of ONCOaccuPanel for Targeted Next-Generation Sequencing of Solid Tumors
Moonsik Kim, Changseon Lee, Juyeon Hong, Juhee Kim, Ji Yun Jeong, Nora Jee-Young Park, Ji-Eun Kim, Ji Young Park
Cancer Research and Treatment.2023; 55(2): 429. CrossRef - Establishing molecular pathology curriculum for pathology trainees and continued medical education: a collaborative work from the Molecular Pathology Study Group of the Korean Society of Pathologists
Jiwon Koh, Ha Young Park, Jeong Mo Bae, Jun Kang, Uiju Cho, Seung Eun Lee, Haeyoun Kang, Min Eui Hong, Jae Kyung Won, Youn-La Choi, Wan-Seop Kim, Ahwon Lee
Journal of Pathology and Translational Medicine.2023; 57(5): 265. CrossRef - Clinical applications of next-generation sequencing in the diagnosis of genetic disorders in Korea: a narrative review
Jihoon G. Yoon, Man Jin Kim, Yong Jin Kwon, Jong-Hee Chae
Journal of the Korean Medical Association.2023; 66(10): 613. CrossRef - Obtaining spatially resolved tumor purity maps using deep multiple instance learning in a pan-cancer study
Mustafa Umit Oner, Jianbin Chen, Egor Revkov, Anne James, Seow Ye Heng, Arife Neslihan Kaya, Jacob Josiah Santiago Alvarez, Angela Takano, Xin Min Cheng, Tony Kiat Hon Lim, Daniel Shao Weng Tan, Weiwei Zhai, Anders Jacobsen Skanderup, Wing-Kin Sung, Hwee
Patterns.2022; 3(2): 100399. CrossRef - Update on Molecular Diagnosis in Extranodal NK/T-Cell Lymphoma and Its Role in the Era of Personalized Medicine
Ka-Hei (Murphy) Sun, Yin-Ting (Heylie) Wong, Ka-Man (Carmen) Cheung, Carmen (Michelle) Yuen, Yun-Tat (Ted) Chan, Wing-Yan (Jennifer) Lai, Chun (David) Chao, Wing-Sum (Katie) Fan, Yuen-Kiu (Karen) Chow, Man-Fai Law, Ho-Chi (Tommy) Tam
Diagnostics.2022; 12(2): 409. CrossRef - Defining Novel DNA Virus-Tumor Associations and Genomic Correlates Using Prospective Clinical Tumor/Normal Matched Sequencing Data
Chad M. Vanderbilt, Anita S. Bowman, Sumit Middha, Kseniya Petrova-Drus, Yi-Wei Tang, Xin Chen, Youxiang Wang, Jason Chang, Natasha Rekhtman, Klaus J. Busam, Sounak Gupta, Meera Hameed, Maria E. Arcila, Marc Ladanyi, Michael F. Berger, Snjezana Dogan, Ahm
The Journal of Molecular Diagnostics.2022; 24(5): 515. CrossRef - Performance Evaluation of Three DNA Sample Tracking Tools in a Whole Exome Sequencing Workflow
Gertjan Wils, Céline Helsmoortel, Pieter-Jan Volders, Inge Vereecke, Mauro Milazzo, Jo Vandesompele, Frauke Coppieters, Kim De Leeneer, Steve Lefever
Molecular Diagnosis & Therapy.2022; 26(4): 411. CrossRef - Clinical Quality Considerations when Using Next-Generation Sequencing (NGS) in Clinical Drug Development
Timothé Ménard, Alaina Barros, Christopher Ganter
Therapeutic Innovation & Regulatory Science.2021; 55(5): 1066. CrossRef - Fast Healthcare Interoperability Resources (FHIR)–Based Quality Information Exchange for Clinical Next-Generation Sequencing Genomic Testing: Implementation Study
Donghyeong Seong, Sungwon Jung, Sungchul Bae, Jongsuk Chung, Dae-Soon Son, Byoung-Kee Yi
Journal of Medical Internet Research.2021; 23(4): e26261. CrossRef - Status of Next-Generation Sequencing-Based Genetic Diagnosis in Hematologic Malignancies in Korea (2017-2018)
JinJu Kim, Ja Young Lee, Jungwon Huh, Myung-Hyun Nam, Myungshin Kim, Young-Uk Cho, Sun-Young Kong, Seung-Tae Lee, In-Suk Kim
Laboratory Medicine Online.2021; 11(1): 25. CrossRef - MSI-Testung
Josef Rüschoff, Gustavo Baretton, Hendrik Bläker, Wolfgang Dietmaier, Manfred Dietel, Arndt Hartmann, Lars-Christian Horn, Korinna Jöhrens, Thomas Kirchner, Ruth Knüchel, Doris Mayr, Sabine Merkelbach-Bruse, Hans-Ulrich Schildhaus, Peter Schirmacher, Mark
Der Pathologe.2021; 42(4): 414. CrossRef - Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
Journal of Pathology and Translational Medicine.2021; 55(3): 181. CrossRef - MSI testing
Josef Rüschoff, Gustavo Baretton, Hendrik Bläker, Wolfgang Dietmaier, Manfred Dietel, Arndt Hartmann, Lars-Christian Horn, Korinna Jöhrens, Thomas Kirchner, Ruth Knüchel, Doris Mayr, Sabine Merkelbach-Bruse, Hans-Ulrich Schildhaus, Peter Schirmacher, Mark
Der Pathologe.2021; 42(S1): 110. CrossRef - 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer
Huisong Lee, Hyeon Kook Lee, Seog Ki Min, Won Hee Lee
World Journal of Surgical Oncology.2020;[Epub] CrossRef - Risk Stratification Using a Novel Genetic Classifier IncludingPLEKHS1Promoter Mutations for Differentiated Thyroid Cancer with Distant Metastasis
Chan Kwon Jung, Seung-Hyun Jung, Sora Jeon, Young Mun Jeong, Yourha Kim, Sohee Lee, Ja-Seong Bae, Yeun-Jun Chung
Thyroid.2020; 30(11): 1589. CrossRef - Biomarker testing for advanced lung cancer by next-generation sequencing; a valid method to achieve a comprehensive glimpse at mutational landscape
Anurag Mehta, Smreti Vasudevan, Sanjeev Kumar Sharma, Manoj Panigrahi, Moushumi Suryavanshi, Mumtaz Saifi, Ullas Batra
Applied Cancer Research.2020;[Epub] CrossRef - Application Areas of Traditional Molecular Genetic Methods and NGS in relation to Hereditary Urological Cancer Diagnosis
Dmitry S. Mikhaylenko, Alexander S. Tanas, Dmitry V. Zaletaev, Marina V. Nemtsova
Journal of Oncology.2020; 2020: 1. CrossRef - Assembling and Validating Bioinformatic Pipelines for Next-Generation Sequencing Clinical Assays
Jeffrey A SoRelle, Megan Wachsmann, Brandi L. Cantarel
Archives of Pathology & Laboratory Medicine.2020; 144(9): 1118. CrossRef - Standard operating procedure for somatic variant refinement of sequencing data with paired tumor and normal samples
Erica K. Barnell, Peter Ronning, Katie M. Campbell, Kilannin Krysiak, Benjamin J. Ainscough, Lana M. Sheta, Shahil P. Pema, Alina D. Schmidt, Megan Richters, Kelsy C. Cotto, Arpad M. Danos, Cody Ramirez, Zachary L. Skidmore, Nicholas C. Spies, Jasreet Hun
Genetics in Medicine.2019; 21(4): 972. CrossRef - A DNA pool of FLT3-ITD positive DNA samples can be used efficiently for analytical evaluation of NGS-based FLT3-ITD quantitation - Testing several different ITD sequences and rates, simultaneously
Zoltán A. Mezei, Dávid Tornai, Róza Földesi, László Madar, Andrea Sümegi, Mária Papp, Péter Antal-Szalmás
Journal of Biotechnology.2019; 303: 25. CrossRef - Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges
Catriona Hippman, Corey Nislow
Journal of Personalized Medicine.2019; 9(3): 40. CrossRef - Cancer Panel Assay for Precision Oncology Clinic: Results from a 1-Year Study
Dohee Kwon, Binnari Kim, Hyeong Chan Shin, Eun Ji Kim, Sang Yun Ha, Kee-Taek Jang, Seung Tae Kim, Jeeyun Lee, Won Ki Kang, Joon Oh Park, Kyoung-Mee Kim
Translational Oncology.2019; 12(11): 1488. CrossRef - Analytical Evaluation of an NGS Testing Method for Routine Molecular Diagnostics on Melanoma Formalin-Fixed, Paraffin-Embedded Tumor-Derived DNA
Irene Mancini, Lisa Simi, Francesca Salvianti, Francesca Castiglione, Gemma Sonnati, Pamela Pinzani
Diagnostics.2019; 9(3): 117. CrossRef - Benchmark Database for Process Optimization and Quality Control of Clinical Cancer Panel Sequencing
Donghyeong Seong, Jongsuk Chung, Ki-Wook Lee, Sook-Young Kim, Byung-Suk Kim, Jung-Keun Song, Sungwon Jung, Taeseob Lee, Donghyun Park, Byoung-Kee Yi, Woong-Yang Park, Dae-Soon Son
Biotechnology and Bioprocess Engineering.2019; 24(5): 793. CrossRef - Use of the Ion PGM and the GeneReader NGS Systems in Daily Routine Practice for Advanced Lung Adenocarcinoma Patients: A Practical Point of View Reporting a Comparative Study and Assessment of 90 Patients
Simon Heeke, Véronique Hofman, Elodie Long-Mira, Virginie Lespinet, Salomé Lalvée, Olivier Bordone, Camille Ribeyre, Virginie Tanga, Jonathan Benzaquen, Sylvie Leroy, Charlotte Cohen, Jérôme Mouroux, Charles Marquette, Marius Ilié, Paul Hofman
Cancers.2018; 10(4): 88. CrossRef - Use of the Ion AmpliSeq Cancer Hotspot Panel in clinical molecular pathology laboratories for analysis of solid tumours: With emphasis on validation with relevant single molecular pathology tests and the Oncomine Focus Assay
Ahwon Lee, Sung-Hak Lee, Chan Kwon Jung, Gyungsin Park, Kyo Young Lee, Hyun Joo Choi, Ki Ouk Min, Tae Jung Kim, Eun Jung Lee, Youn Soo Lee
Pathology - Research and Practice.2018; 214(5): 713. CrossRef - Recent Advancement of the Molecular Diagnosis in Pediatric Brain Tumor
Jeong-Mo Bae, Jae-Kyung Won, Sung-Hye Park
Journal of Korean Neurosurgical Society.2018; 61(3): 376. CrossRef - The long tail of molecular alterations in non-small cell lung cancer: a single-institution experience of next-generation sequencing in clinical molecular diagnostics
Caterina Fumagalli, Davide Vacirca, Alessandra Rappa, Antonio Passaro, Juliana Guarize, Paola Rafaniello Raviele, Filippo de Marinis, Lorenzo Spaggiari, Chiara Casadio, Giuseppe Viale, Massimo Barberis, Elena Guerini-Rocco
Journal of Clinical Pathology.2018; 71(9): 767. CrossRef - Clinical laboratory utilization management and improved healthcare performance
Christopher Naugler, Deirdre L. Church
Critical Reviews in Clinical Laboratory Sciences.2018; 55(8): 535. CrossRef - Development of HLA-A, -B and -DR Typing Method Using Next-Generation Sequencing
Dong Hee Seo, Jeong Min Lee, Mi Ok Park, Hyun Ju Lee, Seo Yoon Moon, Mijin Oh, So Young Kim, Sang-Heon Lee, Ki-Eun Hyeong, Hae-Jin Hu, Dae-Yeon Cho
The Korean Journal of Blood Transfusion.2018; 29(3): 310. CrossRef - Value-based genomics
Jun Gong, Kathy Pan, Marwan Fakih, Sumanta Pal, Ravi Salgia
Oncotarget.2018; 9(21): 15792. CrossRef
- Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis
- KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay
- Sung Hak Lee, Arthur Minwoo Chung, Ahwon Lee, Woo Jin Oh, Yeong Jin Choi, Youn-Soo Lee, Eun Sun Jung
- J Pathol Transl Med. 2017;51(1):24-31. Published online December 25, 2016
- DOI: https://doi.org/10.4132/jptm.2016.10.03
- 12,389 View
- 170 Download
- 5 Web of Science
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Mutations in the KRAS gene have been identified in approximately 50% of colorectal cancers (CRCs). KRAS mutations are well established biomarkers in anti–epidermal growth factor receptor therapy. Therefore, assessment of KRAS mutations is needed in CRC patients to ensure appropriate treatment.
Methods
We compared the analytical performance of the cobas test to Sanger sequencing in 264 CRC cases. In addition, discordant specimens were evaluated by 454 pyrosequencing.
Results
KRAS mutations for codons 12/13 were detected in 43.2% of cases (114/264) by Sanger sequencing. Of 257 evaluable specimens for comparison, KRAS mutations were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% positive percent agreement (PPA) and 91.0% negative percent agreement (NPA) for codons 12/13. Results from the cobas test and Sanger sequencing were discordant for 20 cases (7.8%). Twenty discrepant cases were subsequently subjected to 454 pyrosequencing. After comprehensive analysis of the results from combined Sanger sequencing–454 pyrosequencing and the cobas test, PPA was 97.5% and NPA was 100%.
Conclusions
The cobas test is an accurate and sensitive test for detecting KRAS-activating mutations and has analytical power equivalent to Sanger sequencing. Prescreening using the cobas test with subsequent application of Sanger sequencing is the best strategy for routine detection of KRAS mutations in CRC. -
Citations
Citations to this article as recorded by- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
Journal of Pathology and Translational Medicine.2023; 57(4): 217. CrossRef - Assessment of KRAS and NRAS status in metastatic colorectal cancer: Experience of the National Institute of Oncology in Rabat Morocco
Chaimaa Mounjid, Hajar El Agouri, Youssef Mahdi, Abdelilah Laraqui, En-nacer Chtati, Soumaya Ech-charif, Mouna Khmou, Youssef Bakri, Amine Souadka, Basma El Khannoussi
Annals of Cancer Research and Therapy.2022; 30(2): 80. CrossRef - The current understanding on the impact of KRAS on colorectal cancer
Mingjing Meng, Keying Zhong, Ting Jiang, Zhongqiu Liu, Hiu Yee Kwan, Tao Su
Biomedicine & Pharmacotherapy.2021; 140: 111717. CrossRef - Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer
Barbora Vanova, Michal Kalman, Karin Jasek, Ivana Kasubova, Tatiana Burjanivova, Anna Farkasova, Peter Kruzliak, Dietrich Busselberg, Lukas Plank, Zora Lasabova
Clinical and Experimental Medicine.2019; 19(2): 219. CrossRef - CRISPR Technology for Breast Cancer: Diagnostics, Modeling, and Therapy
Rachel L. Mintz, Madeleine A. Gao, Kahmun Lo, Yeh‐Hsing Lao, Mingqiang Li, Kam W. Leong
Advanced Biosystems.2018;[Epub] CrossRef
- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
- Comparison of Three
BRAF Mutation Tests in Formalin-Fixed Paraffin Embedded Clinical Samples - Soomin Ahn, Jeeyun Lee, Ji-Youn Sung, So Young Kang, Sang Yun Ha, Kee-Taek Jang, Yoon-La Choi, Jung-Sun Kim, Young Lyun Oh, Kyoung-Mee Kim
- Korean J Pathol. 2013;47(4):348-354. Published online August 26, 2013
- DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.4.348
- 9,863 View
- 60 Download
- 9 Crossref
-
 Abstract
Abstract
 PDF
PDF Background Recently,
BRAF inhibitors showed dramatic treatment outcomes inBRAF V600 mutant melanoma. Therefore, the accuracy ofBRAF mutation test is critical.Methods BRAF mutations were tested by dual-priming oligonucleotide-polymerase chain reaction (DPO-PCR), direct sequencing and subsequently retested with a real-time PCR assay, cobas 4800 V600 mutation test. In total, 64 tumors including 34 malignant melanomas and 16 papillary thyroid carcinomas were analyzed. DNA was extracted from formalin-fixed paraffin embedded tissue samples and the results of cobas test were directly compared with those of DPO-PCR and direct sequencing.Results BRAF mutations were found in 23 of 64 (35.9%) tumors. There was 9.4% discordance among 3 methods. Out of 6 discordant cases, 4 cases were melanomas; 3 cases wereBRAF V600E detected only by cobas test, but were not detected by DPO-PCR and direct sequencing. One melanoma patient withBRAF mutation detected only by cobas test has been on vemurafenib treatment for 6 months and showed a dramatic response to vemurafenib. DPO-PCR failed to detect V600K mutation in one case identified by both direct sequencing and cobas test.Conclusions In direct comparison of the currently available DPO-PCR, direct sequencing and real-time cobas test for
BRAF mutation, real-time PCR assay is the most sensitive method.-
Citations
Citations to this article as recorded by- Preoperative BRAFV600E mutation detection in thyroid carcinoma by immunocytochemistry
Kristine Zøylner Swan, Stine Horskær Madsen, Steen Joop Bonnema, Viveque Egsgaard Nielsen, Marie Louise Jespersen
APMIS.2022; 130(11): 627. CrossRef - Strategy to reduce unnecessary surgeries in thyroid nodules with cytology of Bethesda category III (AUS/FLUS): a retrospective analysis of 667 patients diagnosed by surgery
Yong Joon Suh, Yeon Ju Choi
Endocrine.2020; 69(3): 578. CrossRef - A new primer construction technique that effectively increases amplification of rare mutant templates in samples
Jr-Kai Huang, Ling Fan, Tao-Yeuan Wang, Pao-Shu Wu
BMC Biotechnology.2019;[Epub] CrossRef - BRAF and NRAS mutations and antitumor immunity in Korean malignant melanomas and their prognostic relevance: Gene set enrichment analysis and CIBERSORT analysis
Kyueng-Whan Min, Ji-Young Choe, Mi Jung Kwon, Hye Kyung Lee, Ho Suk Kang, Eun Sook Nam, Seong Jin Cho, Hye-Rim Park, Soo Kee Min, Jinwon Seo, Yun Joong Kim, Nan Young Kim, Ho Young Kim
Pathology - Research and Practice.2019; 215(12): 152671. CrossRef - The association between dermoscopic features and BRAF mutational status in cutaneous melanoma: Significance of the blue-white veil
Miquel Armengot-Carbó, Eduardo Nagore, Zaida García-Casado, Rafael Botella-Estrada
Journal of the American Academy of Dermatology.2018; 78(5): 920. CrossRef - Comparison of Five Different Assays for the Detection of BRAF Mutations in Formalin-Fixed Paraffin Embedded Tissues of Patients with Metastatic Melanoma
Claire Franczak, Julia Salleron, Cindy Dubois, Pierre Filhine-Trésarrieu, Agnès Leroux, Jean-Louis Merlin, Alexandre Harlé
Molecular Diagnosis & Therapy.2017; 21(2): 209. CrossRef - Validation of an NGS mutation detection panel for melanoma
Anne Reiman, Hugh Kikuchi, Daniela Scocchia, Peter Smith, Yee Wah Tsang, David Snead, Ian A Cree
BMC Cancer.2017;[Epub] CrossRef - Transformation to Small Cell Lung Cancer of Pulmonary Adenocarcinoma: Clinicopathologic Analysis of Six Cases
Soomin Ahn, Soo Hyun Hwang, Joungho Han, Yoon-La Choi, Se-Hoon Lee, Jin Seok Ahn, Keunchil Park, Myung-Ju Ahn, Woong-Yang Park
Journal of Pathology and Translational Medicine.2016; 50(4): 258. CrossRef - Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma
Katerina Dvorak, Birte Aggeler, John Palting, Penny McKelvie, Andrew Ruszkiewicz, Paul Waring
Pathology.2014; 46(6): 509. CrossRef
- Preoperative BRAFV600E mutation detection in thyroid carcinoma by immunocytochemistry
- Comparison of Direct Sequencing, PNA Clamping-Real Time Polymerase Chain Reaction, and Pyrosequencing Methods for the Detection of
EGFR Mutations in Non-small Cell Lung Carcinoma and the Correlation with Clinical Responses to EGFR Tyrosin - Hyun Ju Lee, Xianhua Xu, Hyojin Kim, Yan Jin, Pingli Sun, Ji Eun Kim, Jin-Haeng Chung
- Korean J Pathol. 2013;47(1):52-60. Published online February 25, 2013
- DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.1.52
- 12,685 View
- 93 Download
- 31 Crossref
-
 Abstract
Abstract
 PDF
PDF Background The aims of this study were to evaluate the abilities of direct sequencing (DS), peptide nucleic acid (PNA) clamping, and pyrosequencing methods to detect epidermal growth factor receptor (
EGFR ) mutations in formalin-fixed paraffin-embedded (FFPE) non-small cell lung carcinoma (NSCLC) samples and to correlateEGFR mutational status as determined by each method with the clinical response toEGFR tyrosine kinase inhibitors (TKIs).Methods Sixty-one NSCLC patients treated with EGFR TKIs were identified to investigate somatic mutations in the
EGFR gene (exons 18-21).Results Mutations in the
EGFR gene were detected in 38 of the 61 patients (62%) by DS, 35 (57%) by PNA clamping and 37 (61%) by pyrosequencing. A total of 44 mutations (72%) were found by at least one of the three methods, and the concordances among the results were relatively high (82-85%; kappa coefficient, 0.713 to 0.736). There were 15 discordant cases (25%) among the three different methods.Conclusions All three
EGFR mutation tests had good concordance rates (over 82%) for FFPE samples. These results suggest that if the DNA quality and enrichment of tumor cells are assured, then DS, PNA clamping, and pyrosequencing are appropriate methods for the detection ofEGFR mutations.-
Citations
Citations to this article as recorded by- Recent Trends of Lung Cancer in Korea
Jae Guk Lee, Ho Cheol Kim, Chang-Min Choi
Tuberculosis and Respiratory Diseases.2021; 84(2): 89. CrossRef - Predictive value of KRAS mutation and excision repair cross-complementing 1 (ERCC1) protein overexpression in patients with colorectal cancer administered FOLFOX regimen
Sun Min Park, Sung Bong Choi, Yoon Suk Lee, In Kyu Lee
Asian Journal of Surgery.2021; 44(5): 715. CrossRef - Recent advances in diagnostic technologies in lung cancer
Hye Jung Park, Sang Hoon Lee, Yoon Soo Chang
The Korean Journal of Internal Medicine.2020; 35(2): 257. CrossRef - Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p.L747P and p.L747S mutations
Sheng-Kai Liang, Jen-Chung Ko, James Chih-Hsin Yang, Jin-Yuan Shih
Lung Cancer.2019; 133: 103. CrossRef - Biomarker Testing for Patients With Advanced Non–Small Cell Lung Cancer: Real-World Issues and Tough Choices
Nathan A. Pennell, Maria E. Arcila, David R. Gandara, Howard West
American Society of Clinical Oncology Educational Book.2019; (39): 531. CrossRef - Evaluation of EGFR mutations in NSCLC with highly sensitive droplet digital PCR assays
Xi‑Wen Jiang, Wei Liu, Xiao‑Ya Zhu, Xiao‑Xie Xu
Molecular Medicine Reports.2019;[Epub] CrossRef - Peptide Nucleic Acid Clamping and Direct Sequencing in the Detection of Oncogenic Alterations in Lung Cancer: Systematic Review and Meta-Analysis
Jae-Uk Song, Jonghoo Lee
Yonsei Medical Journal.2018; 59(2): 211. CrossRef - Distribution of KRAS, DDR2, and TP53 gene mutations in lung cancer: An analysis of Iranian patients
Zahra Fathi, Seyed Ali Javad Mousavi, Raheleh Roudi, Farideh Ghazi, Sumitra Deb
PLOS ONE.2018; 13(7): e0200633. CrossRef - EGFR T790M mutation testing within the osimertinib AURA Phase I study
Simon Dearden, Helen Brown, Suzanne Jenkins, Kenneth S. Thress, Mireille Cantarini, Rebecca Cole, Malcolm Ranson, Pasi A. Jänne
Lung Cancer.2017; 109: 9. CrossRef - Molecular Testing of Lung Cancers
Hyo Sup Shim, Yoon-La Choi, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Mee Sook Roh, Tae-Jung Kim, Seung Yeon Ha, Jin-Haeng Chung, Se Jin Jang, Geon Kook Lee
Journal of Pathology and Translational Medicine.2017; 51(3): 242. CrossRef - Mutations of the Epidermal Growth Factor Receptor Gene in Triple-Negative Breast Cancer
Aeri Kim, Min Hye Jang, Soo Jung Lee, Young Kyung Bae
Journal of Breast Cancer.2017; 20(2): 150. CrossRef - Double primary lung adenocarcinoma diagnosed by epidermal growth factor receptor mutation status
Oh Jung Kwon, Min Hyeok Lee, Sung Ju Kang, Seul Gi Kim, In Beom Jeong, Ji Yun Jeong, Eun Jung Cha, Do Yeun Cho, Young Jin Kim, Ji Woong Son
Yeungnam University Journal of Medicine.2017; 34(2): 270. CrossRef - Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing
Mi-Young Park, Min Hee Jung, Eun Young Eo, Seokjoong Kim, Sang Hoon Lee, Yeon Joo Lee, Jong Sun Park, Young Jae Cho, Jin Haeng Chung, Cheol Hyeon Kim, Ho Il Yoon, Jae Ho Lee, Choon-Taek Lee
Oncotarget.2017; 8(22): 36331. CrossRef - Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study
Kyueng-Whan Min, Wan-Seop Kim, Se Jin Jang, Yoo Duk Choi, Sunhee Chang, Soon Hee Jung, Lucia Kim, Mee Sook Roh, Choong Sik Lee, Jung Weon Shim, Mi Jin Kim, Geon Kook Lee
QJM.2016; 109(3): 167. CrossRef - Epidermal Growth Factor Receptor Mutation and Anaplastic Lymphoma Kinase Gene Fusion: Detection in Malignant Pleural Effusion by RNA or PNA Analysis
Yi-Lin Chen, Chung-Ta Lee, Cheng-Chan Lu, Shu-Ching Yang, Wan-Li Chen, Yang-Cheng Lee, Chung-Hsien Yang, Shu-Ling Peng, Wu-Chou Su, Nan-Haw Chow, Chung-Liang Ho, Javier S Castresana
PLOS ONE.2016; 11(6): e0158125. CrossRef - IDH Mutation Analysis in Ewing Sarcoma Family Tumors
Ki Yong Na, Byeong-Joo Noh, Ji-Youn Sung, Youn Wha Kim, Eduardo Santini Araujo, Yong-Koo Park
Journal of Pathology and Translational Medicine.2015; 49(3): 257. CrossRef - Immunohistochemical demonstration of alteration of β-catenin during tumor metastasis by different mechanisms according to histology in lung cancer
XIANHUA XU, JI EUN KIM, PING-LI SUN, SEOL BONG YOO, HYOJIN KIM, YAN JIN, JIN-HAENG CHUNG
Experimental and Therapeutic Medicine.2015; 9(2): 311. CrossRef - Detection of EGFR-TK Domain–activating Mutations in NSCLC With Generic PCR-based Methods
Rajendra B. Shahi, Sylvia De Brakeleer, Jacques De Grève, Caroline Geers, Peter In’t Veld, Erik Teugels
Applied Immunohistochemistry & Molecular Morphology.2015; 23(3): 163. CrossRef - Frequent aerogenous spread with decreased E-cadherin expression of ROS1- rearranged lung cancer predicts poor disease-free survival
Yan Jin, Ping-Li Sun, Soo Young Park, Hyojin Kim, Eunhyang Park, Gilhyang Kim, Sukki Cho, Kwhanmien Kim, Choon-Taek Lee, Jin-Haeng Chung
Lung Cancer.2015; 89(3): 343. CrossRef - Membranous Insulin-like Growth Factor-1 Receptor (IGF1R) Expression Is Predictive of Poor Prognosis in Patients with Epidermal Growth Factor Receptor (EGFR)-Mutant Lung Adenocarcinoma
Eunhyang Park, Soo Young Park, Hyojin Kim, Ping-Li Sun, Yan Jin, Suk Ki Cho, Kwhanmien Kim, Choon-Taek Lee, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2015; 49(5): 382. CrossRef - Peptide Nucleic Acid Clamping Versus Direct Sequencing for the Detection of EGFR Gene Mutation in Patients with Non-small Cell Lung Cancer
Seong-Hoon Yoon, Yoo-Duk Choi, In-Jae Oh, Kyu-Sik Kim, Hayoung Choi, Jinsun Chang, Hong-Joon Shin, Cheol-Kyu Park, Young-Chul Kim
Cancer Research and Treatment.2015; 47(4): 661. CrossRef - Analysis of Mutations in Epidermal Growth Factor Receptor Gene in Korean Patients with Non-small Cell Lung Cancer: Summary of a Nationwide Survey
Sang Hwa Lee, Wan Seop Kim, Yoo Duk Choi, Jeong Wook Seo, Joung Ho Han, Mi Jin Kim, Lucia Kim, Geon Kook Lee, Chang Hun Lee, Mee Hye Oh, Gou Young Kim, Sun Hee Sung, Kyo Young Lee, Sun Hee Chang, Mee Sook Rho, Han Kyeom Kim, Soon Hee Jung, Se Jin Jang
Journal of Pathology and Translational Medicine.2015; 49(6): 481. CrossRef - Novel EGFR mutation-specific antibodies for lung adenocarcinoma: Highly specific but not sensitive detection of an E746_A750 deletion in exon 19 and an L858R mutation in exon 21 by immunohistochemistry
An Na Seo, Tae-In Park, Yan Jin, Ping-Li Sun, Hyojin Kim, Hyun Chang, Jin-Haeng Chung
Lung Cancer.2014; 83(3): 316. CrossRef - Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non‐small‐cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage
Tae‐Jung Kim, Chan Kwon Park, Chang Dong Yeo, Kihoon Park, Chin Kook Rhee, Jusang Kim, Seung Joon Kim, Sang Haak Lee, Kyo‐Young Lee, Hyoung‐Kyu Yoon
Journal of Surgical Oncology.2014; 110(3): 245. CrossRef - Epidermal growth factor receptor mutations and anaplastic lymphoma kinase rearrangements in lung cancer with nodular ground-glass opacity
Sung-Jun Ko, Yeon Joo Lee, Jong Sun Park, Young-Jae Cho, Ho Il Yoon, Jin-Haeng Chung, Tae Jung Kim, Kyung Won Lee, Kwhanmien Kim, Sanghoon Jheon, Hyojin Kim, Jae Ho Lee, Choon-Taek Lee
BMC Cancer.2014;[Epub] CrossRef - Cytoplasmic YAP Expression is Associated with Prolonged Survival in Patients with Lung Adenocarcinomas and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment
Ping-Li Sun, Ji Eun Kim, Seol Bong Yoo, Hyojin Kim, Yan Jin, Sanghoon Jheon, Kwhanmien Kim, Choon Taek Lee, Jin-Haeng Chung
Annals of Surgical Oncology.2014; 21(S4): 610. CrossRef - Sensitive methods for detection of the S768R substitution in exon 18 of the DDR2 gene in patients with central nervous system metastases of non-small cell lung cancer
Marcin Nicoś, Tomasz Powrózek, Paweł Krawczyk, Bożena Jarosz, Beata Pająk, Marek Sawicki, Krzysztof Kucharczyk, Tomasz Trojanowski, Janusz Milanowski
Medical Oncology.2014;[Epub] CrossRef - Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas
A N Seo, J M Yang, H Kim, S Jheon, K Kim, C T Lee, Y Jin, S Yun, J-H Chung, J H Paik
British Journal of Cancer.2014; 110(11): 2688. CrossRef - KRASMutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing
Boram Lee, Boin Lee, Gangmin Han, Mi Jung Kwon, Joungho Han, Yoon-La Choi
Korean Journal of Pathology.2014; 48(2): 100. CrossRef - Guideline Recommendations forEGFRMutation Testing in Lung Cancer: Proposal of the Korean Cardiopulmonary Pathology Study Group
Hyo Sup Shim, Jin-Haeng Chung, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Geon Kook Lee, Soon-Hee Jung, Se Jin Jang
Korean Journal of Pathology.2013; 47(2): 100. CrossRef - Immunohistochemical Classification of Primary and Secondary Glioblastomas
Kyu Sang Lee, Gheeyoung Choe, Kyung Han Nam, An Na Seo, Sumi Yun, Kyung Ju Kim, Hwa Jin Cho, Sung Hye Park
Korean Journal of Pathology.2013; 47(6): 541. CrossRef
- Recent Trends of Lung Cancer in Korea
- MGMT Gene Promoter Methylation Analysis by Pyrosequencing of Brain Tumour.
- Young Zoon Kim, Young Jin Song, Ki Uk Kim, Dae Cheol Kim
- Korean J Pathol. 2011;45(5):455-462.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.5.455
- 4,718 View
- 16 Download
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
The aim of this study was to determine whether pyrosequencing (PSQ) might be useful to achieve O6-methyl guanine methyltransferase (MGMT) promoter methylation using 1- to 13-year-old archival tissues as a clinical biomarker in routine practice.
METHODS
The study included 141 formalin-fixed paraffin-embedded (FFPE) glial tumors from the archives of the Pathology Department from 1997-2010.
RESULTS
The average percentage of methylation (MP) of the 141 cases was 14.0+/-16.8%, and methylated cases were 32.3+/-14.9%. The average MP of each year did not show a linear increasing or decreasing pattern according to the age of the FFPE block (p=0.771). The average MP of methylated glioblastomas was 35.8+/-14.7%, 31.8+/-15.5% for anaplastic astrocytomas, and 22.4+/-15.1% for astrocytoma. A tendency was observed toward an increasing pattern of average MP with World Health Organization (WHO) grade (p=0.063) in astrocytic tumors. A correlation was observed between average MP and WHO grade (p=0.038) and a bimodal distribution was observed between the methylated and unmethylated cases, using a 9% cut-off value (p<0.001).
CONCLUSIONS
The results showed that a quantitative approach for MGMT promoter methylation yielded a 100% success rate for FFPE tissues from archives. PSQ can be used in a retrospective trial, but the cut-off value and calculation method should be further validated. -
Citations
Citations to this article as recorded by- CDKN2A Homozygous Deletion Is a Stronger Predictor of Outcome than IDH1/2-Mutation in CNS WHO Grade 4 Gliomas
Sang Hyuk Lee, Tae Gyu Kim, Kyeong Hwa Ryu, Seok Hyun Kim, Young Zoon Kim
Biomedicines.2024; 12(10): 2256. CrossRef - Immunohistochemical Classification of Primary and Secondary Glioblastomas
Kyu Sang Lee, Gheeyoung Choe, Kyung Han Nam, An Na Seo, Sumi Yun, Kyung Ju Kim, Hwa Jin Cho, Sung Hye Park
Korean Journal of Pathology.2013; 47(6): 541. CrossRef
- CDKN2A Homozygous Deletion Is a Stronger Predictor of Outcome than IDH1/2-Mutation in CNS WHO Grade 4 Gliomas
- Rapid and Sensitive Detection of KRAS Mutation by Peptide Nucleic Acid-based Real-time PCR Clamping: A Comparison with Direct Sequencing between Fresh Tissue and Formalin-fixed and Paraffin Embedded Tissue of Colorectal Cancer.
- Dongjun Jeong, Yujun Jeong, Jonghyun Lee, Moo Jun Baek, Yongbae Kim, Ji Hye Lee, Hyun Deuk Cho, Mee Hye Oh, Chang Jin Kim
- Korean J Pathol. 2011;45(2):151-159.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.2.151
- 6,040 View
- 84 Download
- 8 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Rapid and sensitive detection of KRAS mutation is needed to maximize the benefits for patients who are being treated with monoclonal antibodies to target the epidermal growth factor receptor in colorectal cancer. The aim of this study is to evaluate the efficacy of the peptide nucleic acid clamp real-time PCR (PCqPCR) as compared to that of direct sequencing (DS) between using fresh colorectal cancer tissue and the matched formalin-fixed and paraffin-embedded (FFPE) colorectal cancer tissue.
METHODS
The efficacy of PCqPCR was evaluated and compared with that of DS using fresh tissue and matched FFPE tissue from 30 cases of colorectal cancer.
RESULTS
PCqPCR is more sensitive than DS for detecting KRAS mutation. PCqPCR detected 1% of mutants in 1 ng DNA. PCqPCR detected mutation in 1% of mutant cells, while DS barely detected, by manual reading, that in 20-50% of mutant cells. In the clinical samples, PCqPCR detected KRAS mutation in 60.0% while DS detected KRAS mutation in 53.3% of the colorectal cancers. The two methods showed a 100% concordance rate for detecting KRAS mutation between the fresh tissue and FFPE tissue.
CONCLUSIONS
The PCqPCR method is efficiently applicable for the detection of KRAS mutation in a clinical setting. -
Citations
Citations to this article as recorded by- NTRK oncogenic fusions are exclusively associated with the serrated neoplasia pathway in the colorectum and begin to occur in sessile serrated lesions
Jung Ho Kim, Jeong Hoon Hong, Yoon‐La Choi, Ji Ae Lee, Mi‐kyoung Seo, Mi‐Sook Lee, Sung Bin An, Min Jung Sung, Nam‐Yun Cho, Sung‐Su Kim, Young Kee Shin, Sangwoo Kim, Gyeong Hoon Kang
The Journal of Pathology.2021; 255(4): 399. CrossRef - Discrimination of the V600E Mutation in BRAF by Rolling Circle Amplification and Förster Resonance Energy Transfer
Mariia Dekaliuk, Xue Qiu, Frédéric Troalen, Pierre Busson, Niko Hildebrandt
ACS Sensors.2019; 4(10): 2786. CrossRef - Sensitive Genotyping of Somatic Mutations in the EGFR, KRAS, PIK3CA, BRAF Genes from NSCLC Patients Using Hydrogel Biochips
Marina Emelyanova, Ksenia Arkhipova, Natalia Mazurenko, Alexander Chudinov, Irina Demidova, Irina Zborovskaya, Lyudmila Lyubchenko, Alexander Zasedatelev, Tatiana Nasedkina
Applied Immunohistochemistry & Molecular Morphology.2015; 23(4): 255. CrossRef - Low Frequency of KRAS Mutation in Pancreatic Ductal Adenocarcinomas in Korean Patients and Its Prognostic Value
Mi Jung Kwon, Jang Yong Jeon, Hye-Rim Park, Eun Sook Nam, Seong Jin Cho, Hyung Sik Shin, Ji Hyun Kwon, Joo Seop Kim, Boram Han, Dong Hoon Kim, Yoon-La Choi
Pancreas.2015; 44(3): 484. CrossRef - Comparison of PNA clamping and direct sequencing for detecting KRAS mutations in matched tumour tissue, cell block, pleural effusion and serum from patients with malignant pleural effusion
Ji Young Kang, Chan Kwon Park, Chang Dong Yeo, Hea Yeon Lee, Chin Kook Rhee, Seung Joon Kim, Seok Chan Kim, Young Kyoon Kim, Mi Sun Park, Hyeon Woo Yim
Respirology.2015; 20(1): 138. CrossRef - BRAF V600E Mutation Analysis in Papillary Thyroid Carcinomas by Peptide Nucleic Acid Clamp Real-time PCR
Dongjun Jeong, Yujun Jeong, Ji Hye Park, Sun Wook Han, Sung Yong Kim, Yeo Joo Kim, Sang Jin Kim, Young Hwangbo, Soyoung Park, Hyun Deuk Cho, Mee Hye Oh, Seung Ha Yang, Chang Jin Kim
Annals of Surgical Oncology.2013; 20(3): 759. CrossRef - Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing
Chang Dong Yeo, Jin Woo Kim, Kwan Hyoung Kim, Jick Hwan Ha, Chin Kook Rhee, Seung Joon Kim, Young Kyoon Kim, Chan Kwon Park, Sang Haak Lee, Mi Sun Park, Hyeon Woo Yim
Lung Cancer.2013; 81(2): 207. CrossRef - Detection ofBRAFV600EMutations in Papillary Thyroid Carcinomas by Peptide Nucleic Acid Clamp Real-Time PCR: A Comparison with Direct Sequencing
Dongjun Jeong, Yujun Jeong, Sungche Lee, Hyeran Lee, Wanju Lee, Hyungjoo Kim, Doosan Park, Soyoung Park, Wenxia Mu, Hyun-Deuk Cho, Mee-Hye Oh, Sung Soo Lee, Seung-Ha Yang, Chang-Jin Kim
Korean Journal of Pathology.2012; 46(1): 61. CrossRef
- NTRK oncogenic fusions are exclusively associated with the serrated neoplasia pathway in the colorectum and begin to occur in sessile serrated lesions
- A Consideration of MGMT Gene Promotor Methylation Analysis for Glioblastoma Using Methylation-Specific Polymerase Chain Reaction and Pyrosequencing.
- Sang Hwa Lee, Tae Sook Hwang, Young Cho Koh, Wook Youn Kim, Hye Seung Han, Wan Seop Kim, Young Sin Ko, So Dug Lim
- Korean J Pathol. 2011;45(1):21-29.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.1.21
- 5,104 View
- 52 Download
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
O6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation is currently the most promising predictive marker for the outcome and benefit from temozolomide treatment in patients with glioblastoma, but there is no consensus on the analysis method for assessing the methylation status in the molecular diagnostic field. The objective of this study was to evaluate methylation-specific polymerase chain reaction (MSP) and pyrosequencing methods for assessing MGMT gene promotor methylation of glioblastoma as well as assessing the MGMT protein expression by immunohistochemistry.
METHODS
Twenty-seven cases of glioblastoma from the archives at the Department of Pathology Konkuk University Hospital were selected. MGMT promoter methylation was evaluated by MSP and the pyrosequencing methods. The MGMT expression was also measured at the protein level by immunohistochemistry.
RESULTS
Overall, MGMT hypermethylation was observed in 44.4% (12/27 cases) of the case of glioblastoma using either MSP or pyrosequencing. The concordant rate was 70.3% (19/27 cases) between MSP and pyrosequencing for MGMT methylation. There was no correlation between MGMT methylation and the protein expression. No significant differences in progression free survival and overall survival were seen between the methylated group and the unmethylated group by using either MSP or pyrosequencing. The status of the MGMT protein expression was correlated with progression free survival (p=0.026).
CONCLUSIONS
In this study the concordance rate between MSP and the pyrosequencing methods for assessing MGMT gene promotor methylation was relatively low for the cases of glioblastoma. This suggests that more reliable techniques for routine MGMT methylation study of glioblastoma remain to be developed because of quality control and assurance issues. -
Citations
Citations to this article as recorded by- Prognostic Role of Methylation Status of theMGMTPromoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients
Dae Cheol Kim, Ki Uk Kim, Young Zoon Kim
Journal of Korean Neurosurgical Society.2016; 59(1): 26. CrossRef - Distinct genetic alterations in pediatric glioblastomas
Sun-ju Byeon, Jae Kyung Myung, Se Hoon Kim, Seung-Ki Kim, Ji Hoon Phi, Sung-Hye Park
Child's Nervous System.2012; 28(7): 1025. CrossRef - MGMTGene Promoter Methylation Analysis by Pyrosequencing of Brain Tumour
Young Zoon Kim, Young Jin Song, Ki Uk Kim, Dae Cheol Kim
The Korean Journal of Pathology.2011; 45(5): 455. CrossRef
- Prognostic Role of Methylation Status of theMGMTPromoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients
- DNA Sequencing of p53 Gene Mutation in Colorectal Carcinomas.
- Young Ran Shim, Joon Hyuk Choi, Won Hee Choi
- Korean J Pathol. 1999;33(6):422-433.
- 2,283 View
- 16 Download
-
 Abstract
Abstract
 PDF
PDF - Mutations in the p53 gene occur during the development of colorectal carcinomas, and play an important role in the conversion of adenoma into carcinoma. To detect the p53 gene mutation and its pattern of expression in colorectal carcinomas, polymerase chain reaction for exons 5, 6, 7, and 8, recombinant gene cloning, and automated DNA sequencing were performed with 30 fresh colorectal carcinomas. Each tissue was also analyzed by immunohistochemical staining for p53 protein. p53 protein was detected in 25 of 30 (83.3%) colorectal carcinomas by immunohistochemical study. p53 mutation was detected in 4 of 30 (13.3%) colorectal carcinomas. The distribution of these mutations among these exons investigated was as follows: Three mutations in exon 5 (66.7%) and 1 mutation in exon 7 (33.3%). One case with mutation in exon 5 had mutations at three different codons. Mutations in exon 5 were found at codon 153 (GGG to AGG: Gly to Arg), 170 (TGC to GGC: Cys to Gly), 186 (CTA to TTA: silent mutation), 158 (GCG to ACG: Ala to Thr), and 176 (ACG to ATG: Thr to Met). Mutation in exon 7 was found at codon 248 (AGG to AGA: silent mutation). Four of them were missense mutations. Two of 6 mutations were silent mutations. Five transition mutations and 1 transversion mutation were also detected. All cases with mutations by automated DNA sequencing showed positive p53 protein immunohistochemical stainining. In conclusion, p53 gene mutation was detected in 4 of 30 (13.3%) colorectal carcinomas, located in codon 153, 158, 170, 176, and 186 of exon 5 and codon 248 of exon 7. Further studies are needed to evaluate the significance of the codon 153 mutation which was not recognized in other studies on colorectal carcinomas.

 E-submission
E-submission
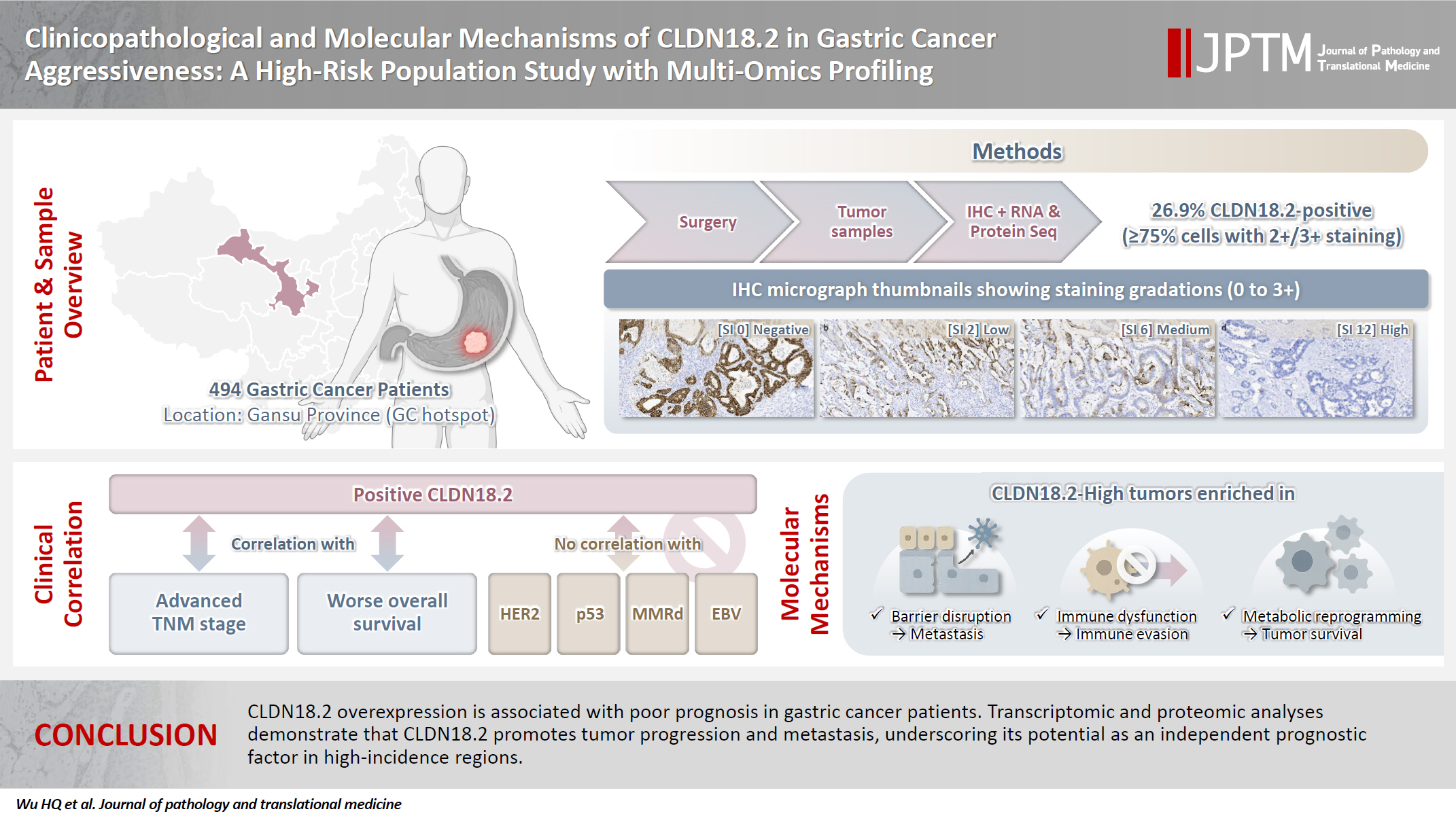
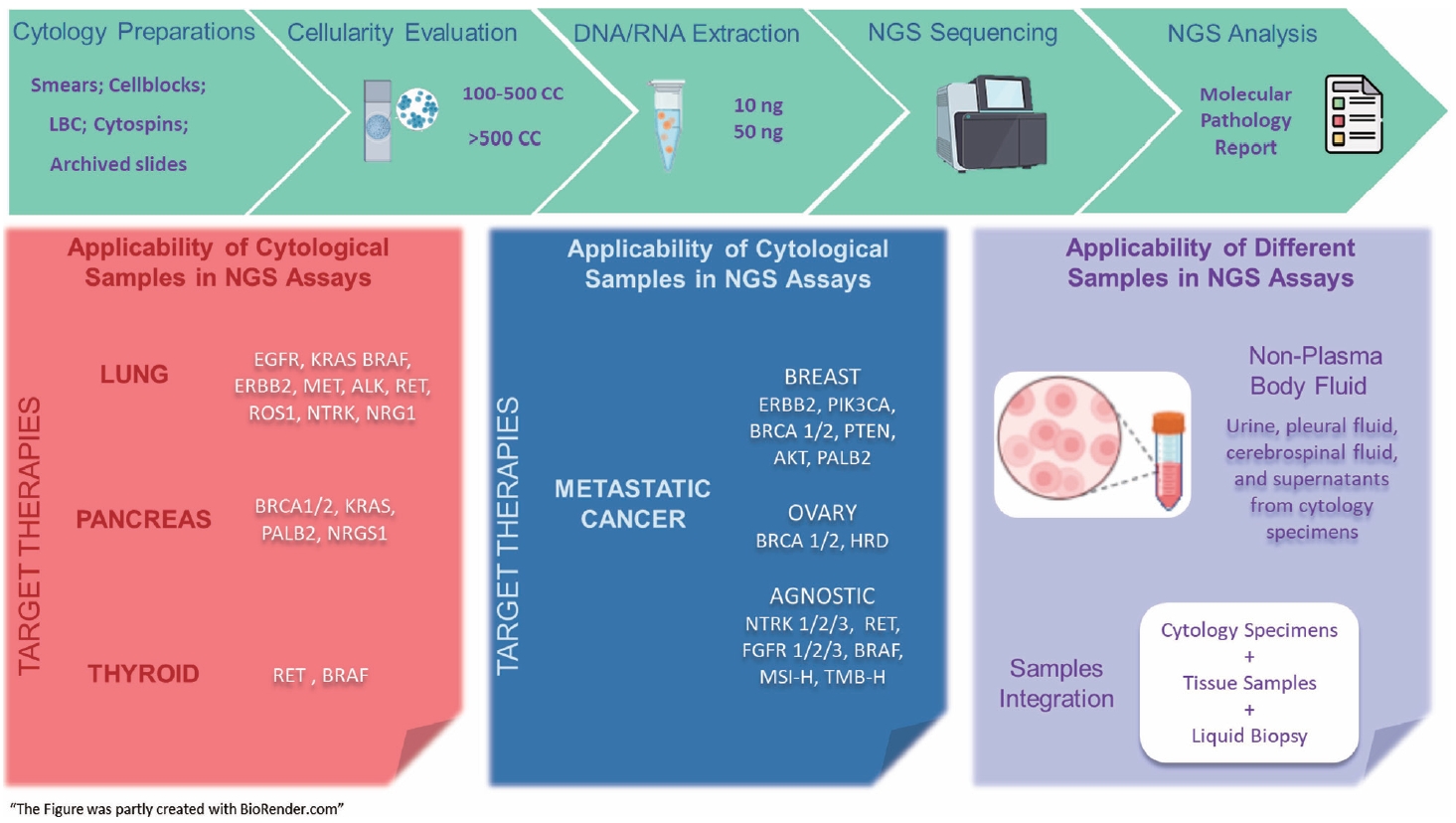

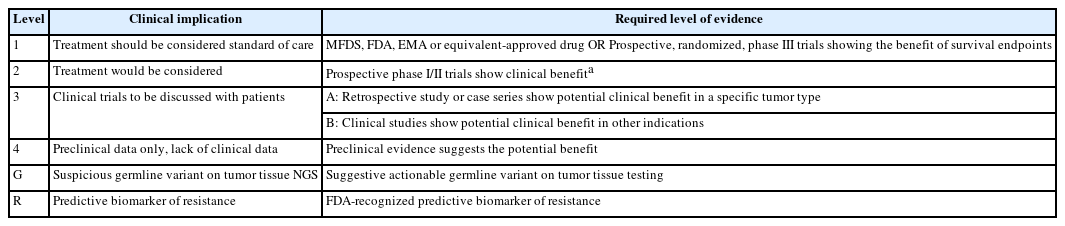
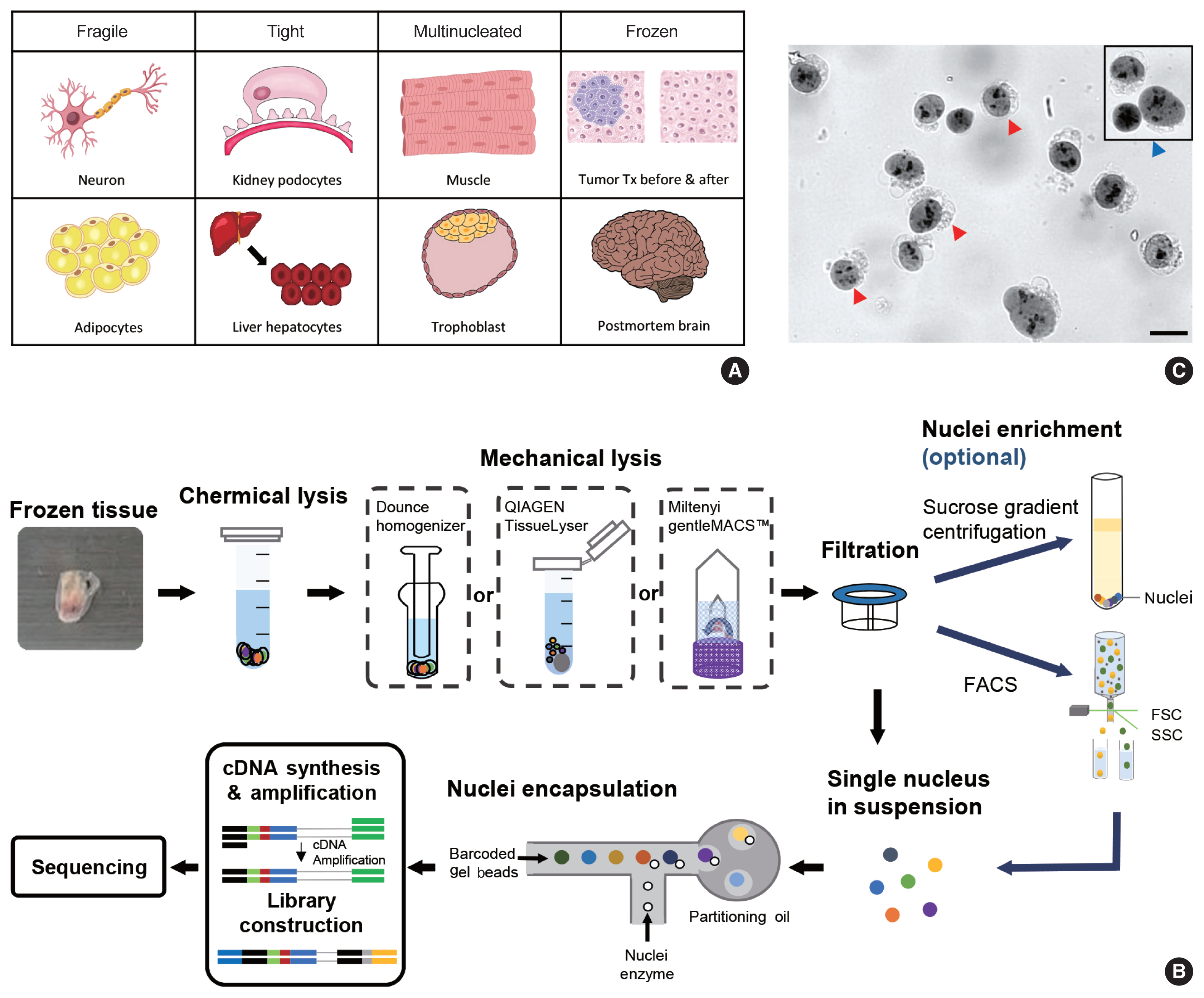
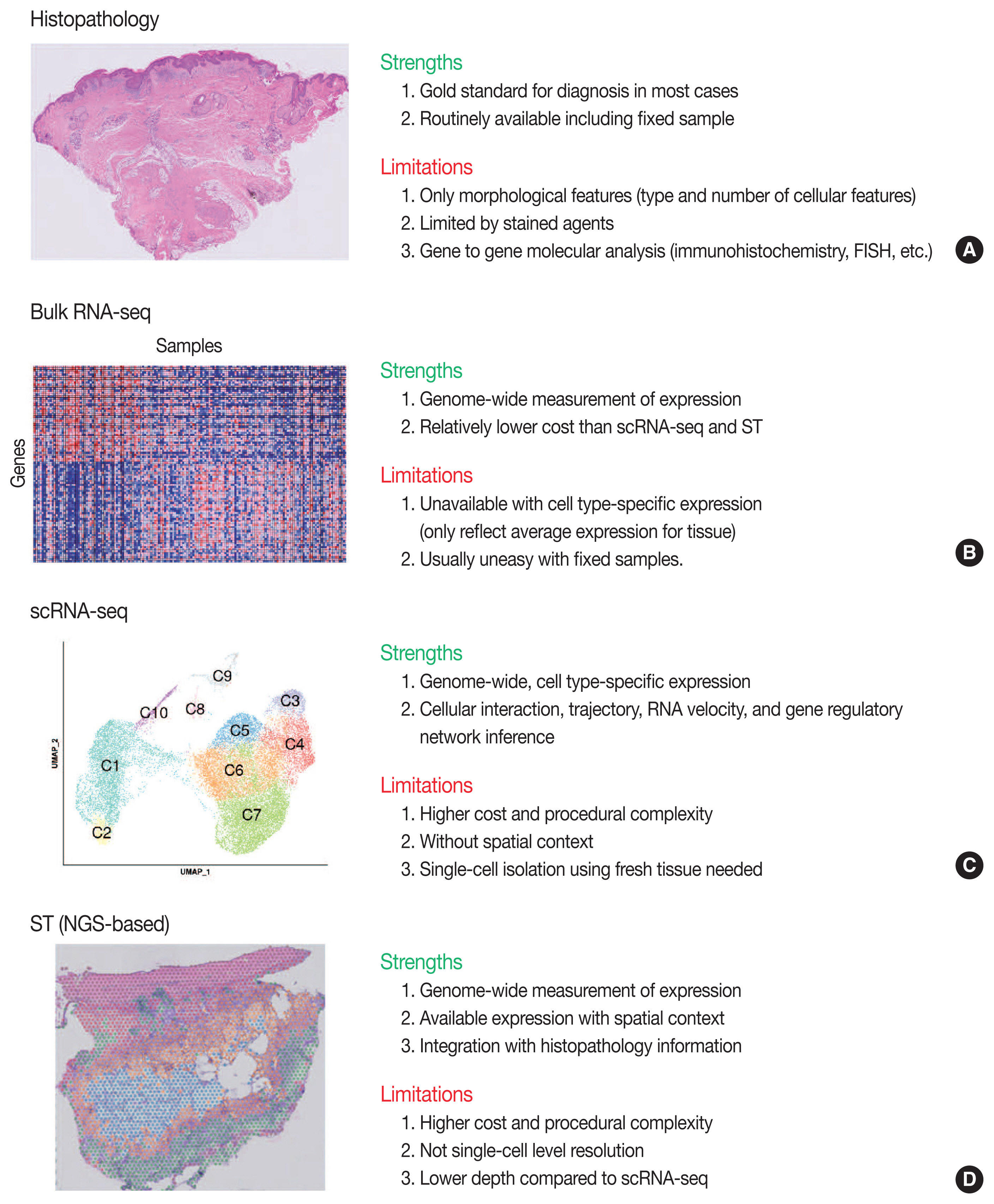
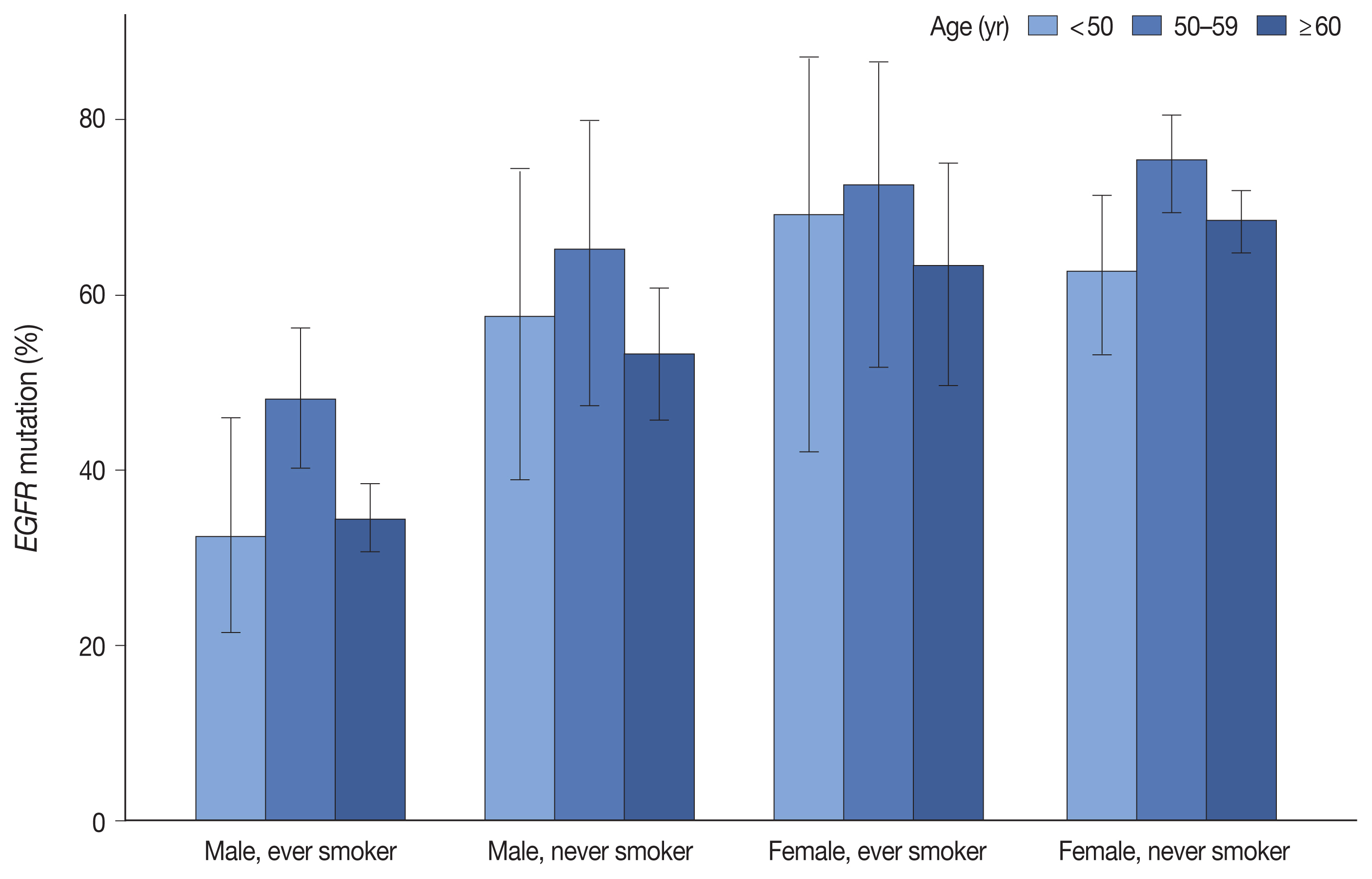
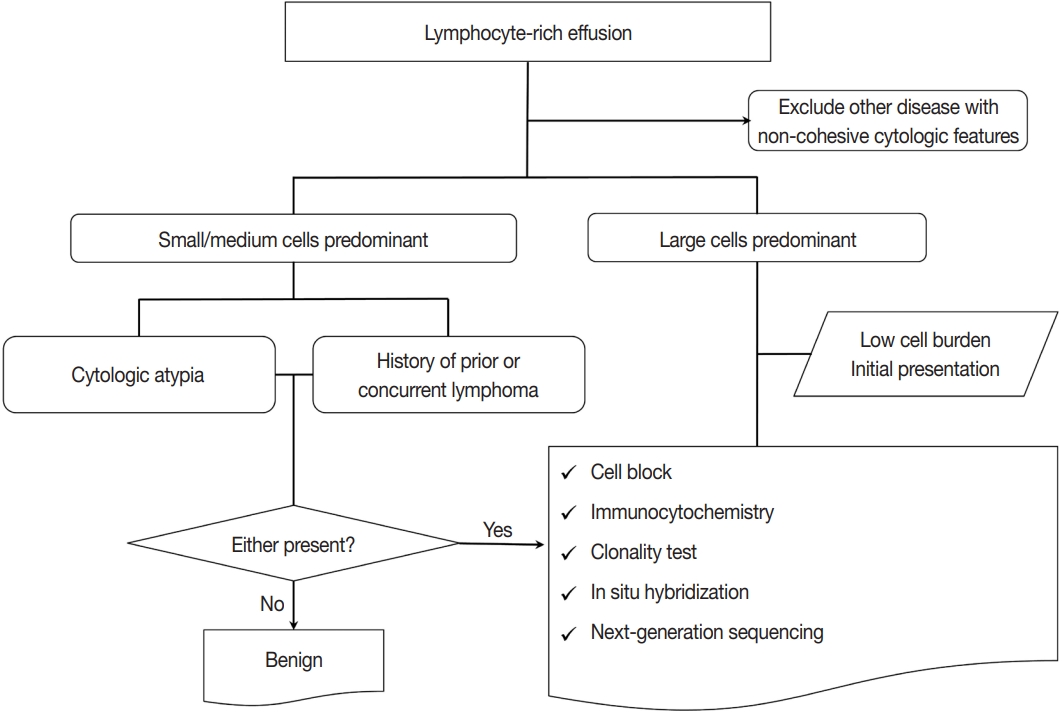

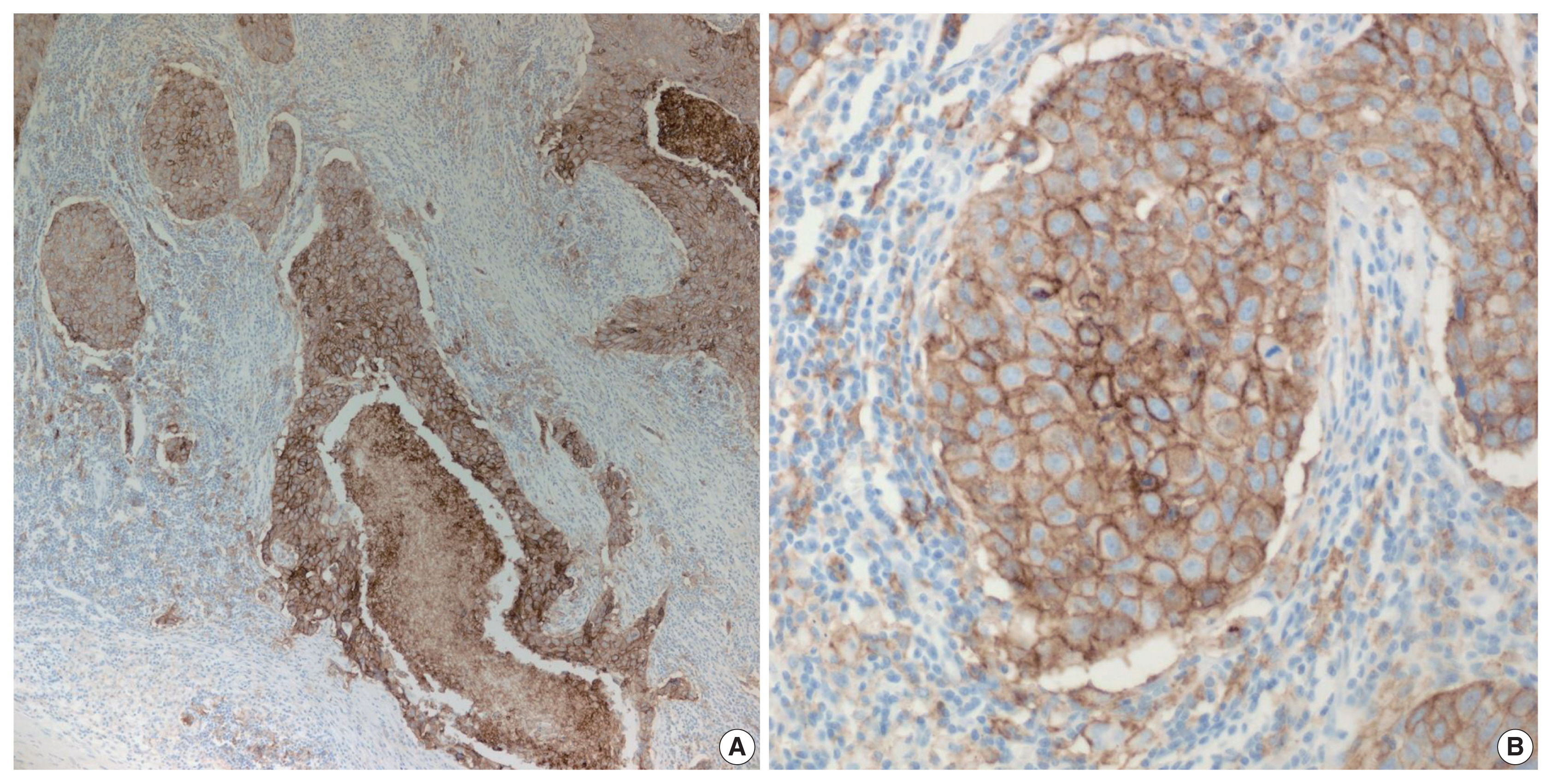
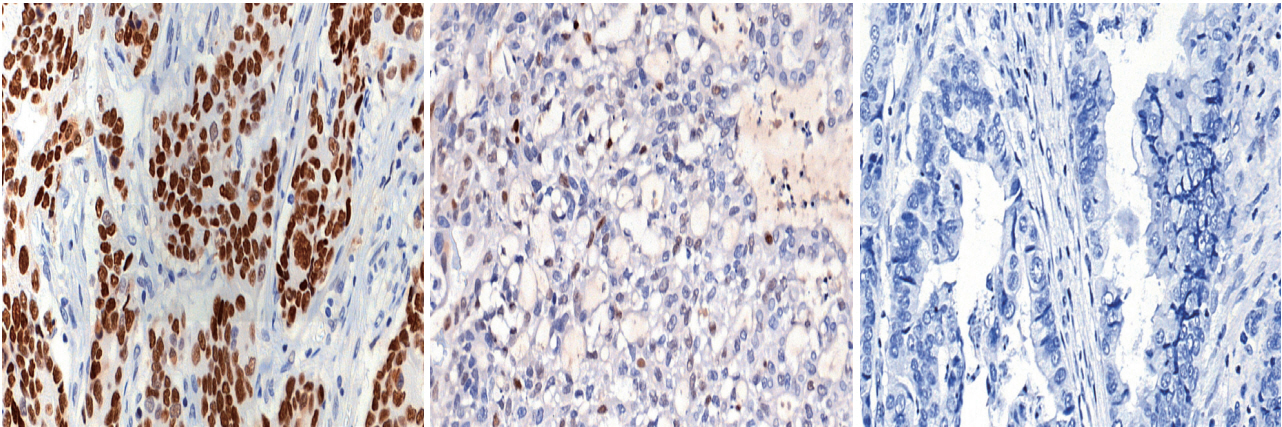
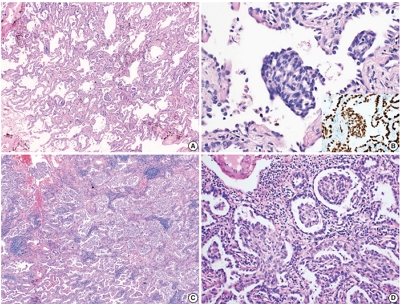
 First
First Prev
Prev



