Search
- Page Path
- HOME > Search
Original Article
- Fatty acid synthetase expression in triple-negative breast cancer
- Jin Hee Park, Hye Seung Han, So Dug Lim, Wook Youn Kim, Kyoung Sik Park, Young Bum Yoo, Seung Eun Lee, Wan-Seop Kim
- J Pathol Transl Med. 2022;56(2):73-80. Published online January 21, 2022
- DOI: https://doi.org/10.4132/jptm.2021.10.27
- 5,689 View
- 201 Download
- 8 Web of Science
- 9 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
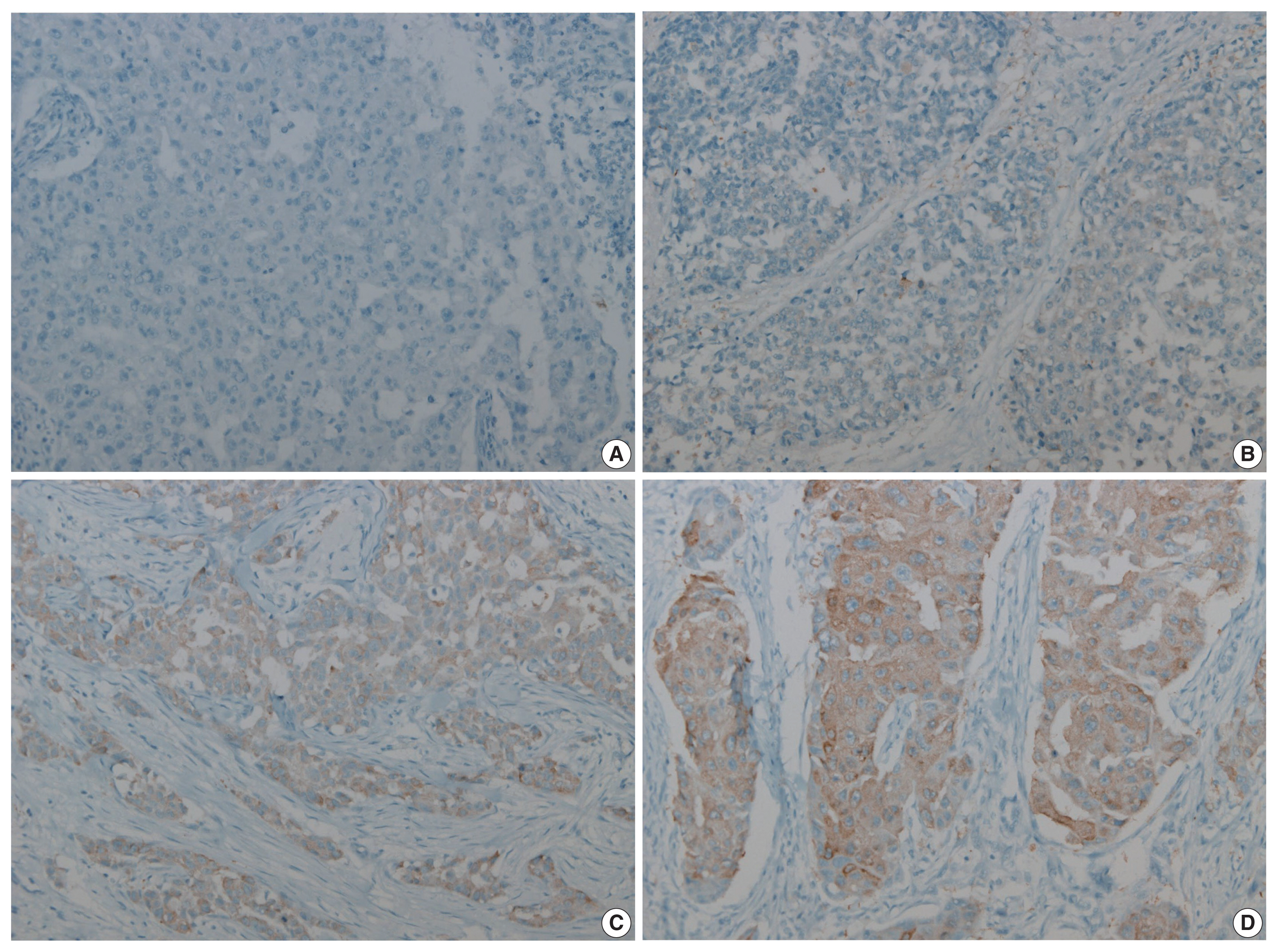
Triple-negative breast cancer (TNBC) has a relatively poor prognosis. Research has identified potential metabolic targets, including fatty acid metabolism, in TNBC. The absence of effective target therapies for TNBC led to exploration of the role of fatty acid synthetase (FASN) as a potential target for TNBC therapy. Here, we analyzed the expression of FASN, a representative lipid metabolism–related protein, and investigated the association between FASN expression and Ki-67 and the programmed death ligand 1 (PD-L1) biomarkers in TNBC.
Methods
Immunohistochemical expression of FASN was analyzed in 166 patients with TNBC. For analytical purposes, patients with 0–1+ FASN staining were grouped as low-grade FASN and patients with 2–3+ FASN staining as high-grade FASN.
Results
FASN expression was observed in 47.1% of TNBC patients. Low and high expression of FASN was identified in 75.9% and 24.1%, respectively, and no statistically significant difference was found in T category, N category, American Joint Committee on Cancer stage, or recurrence rate between the low and high-FASN expression groups. Ki-67 proliferation level was significantly different between the low and high-FASN expression groups. FASN expression was significantly related to Ki-67 as the level increased. There was no significant difference in PD-L1 positivity between the low- and high-FASN expression groups.
Conclusions
We identified FASN expression in 166 TNBC patients. The Ki-67 proliferation index was positively correlated with FASN level, indicating higher proliferation activity as FASN increases. However, there was no statistical association with PD-L1 SP142, the currently FDA-approved assay, or FASN expression level. -
Citations
Citations to this article as recorded by- Lipid metabolism involved in progression and drug resistance of breast cancer
Wenxiang Fu, Aijun Sun, Huijuan Dai
Genes & Diseases.2025; 12(4): 101376. CrossRef - Unveiling the impact of lipid metabolism on triple-negative breast cancer growth and treatment options
Xin-xian Cai, Zhe-zhong Zhang, Xiao-xiao Yang, Wen-rui Shen, Liu-wei Yuan, Xi Ding, Ying Yu, Wen-yu Cai
Frontiers in Oncology.2025;[Epub] CrossRef - Protein biomarkers for diagnosis of breast cancer
Emeka Eze Joshua Iweala, Doris Nnenna Amuji, Faith Chinasaokwu Nnaji
Scientific African.2024; 25: e02308. CrossRef - Microarray analysis points to LMNB1 and JUN as potential target genes for predicting metastasis promotion by etoposide in colorectal cancer
Jiafei Liu, Hongjie Yang, Peng Li, Yuanda Zhou, Zhichun Zhang, Qingsheng Zeng, Xipeng Zhang, Yi Sun
Scientific Reports.2024;[Epub] CrossRef - The signature of extracellular vesicles in hypoxic breast cancer and their therapeutic engineering
Baiheng Zhu, Kehao Xiang, Tanghua Li, Xin Li, Fujun Shi
Cell Communication and Signaling.2024;[Epub] CrossRef - NFYA promotes malignant behavior of triple-negative breast cancer in mice through the regulation of lipid metabolism
Nobuhiro Okada, Chihiro Ueki, Masahiro Shimazaki, Goki Tsujimoto, Susumu Kohno, Hayato Muranaka, Kiyotsugu Yoshikawa, Chiaki Takahashi
Communications Biology.2023;[Epub] CrossRef - Role of EGFR and FASN in breast cancer progression
Suchi Chaturvedi, Mainak Biswas, Sushabhan Sadhukhan, Avinash Sonawane
Journal of Cell Communication and Signaling.2023; 17(4): 1249. CrossRef - Bioinformatics Method Was Used to Analyze the Highly Expressed Gene FAM83A of Breast Cancer in Young Women
Yongzhe Tang, Hao Wang, Qi He, Yuanyuan Chen, Jie Wang, Fahd Abd Algalil
Applied Bionics and Biomechanics.2022; 2022: 1. CrossRef - NCAPH promotes proliferation as well as motility of breast cancer cells by activating the PI3K/AKT pathway
Ting Zhang, Peng Li, Wanying Guo, Qipeng Liu, Weiqiang Qiao, Miao Deng
Physiology International.2022;[Epub] CrossRef
- Lipid metabolism involved in progression and drug resistance of breast cancer
Review
- Programmed cell death-ligand 1 assessment in urothelial carcinoma: prospect and limitation
- Kyu Sang Lee, Gheeyoung Choe
- J Pathol Transl Med. 2021;55(3):163-170. Published online April 7, 2021
- DOI: https://doi.org/10.4132/jptm.2021.02.22
- 4,814 View
- 163 Download
- 6 Web of Science
- 6 Crossref
-
 Abstract
Abstract
 PDF
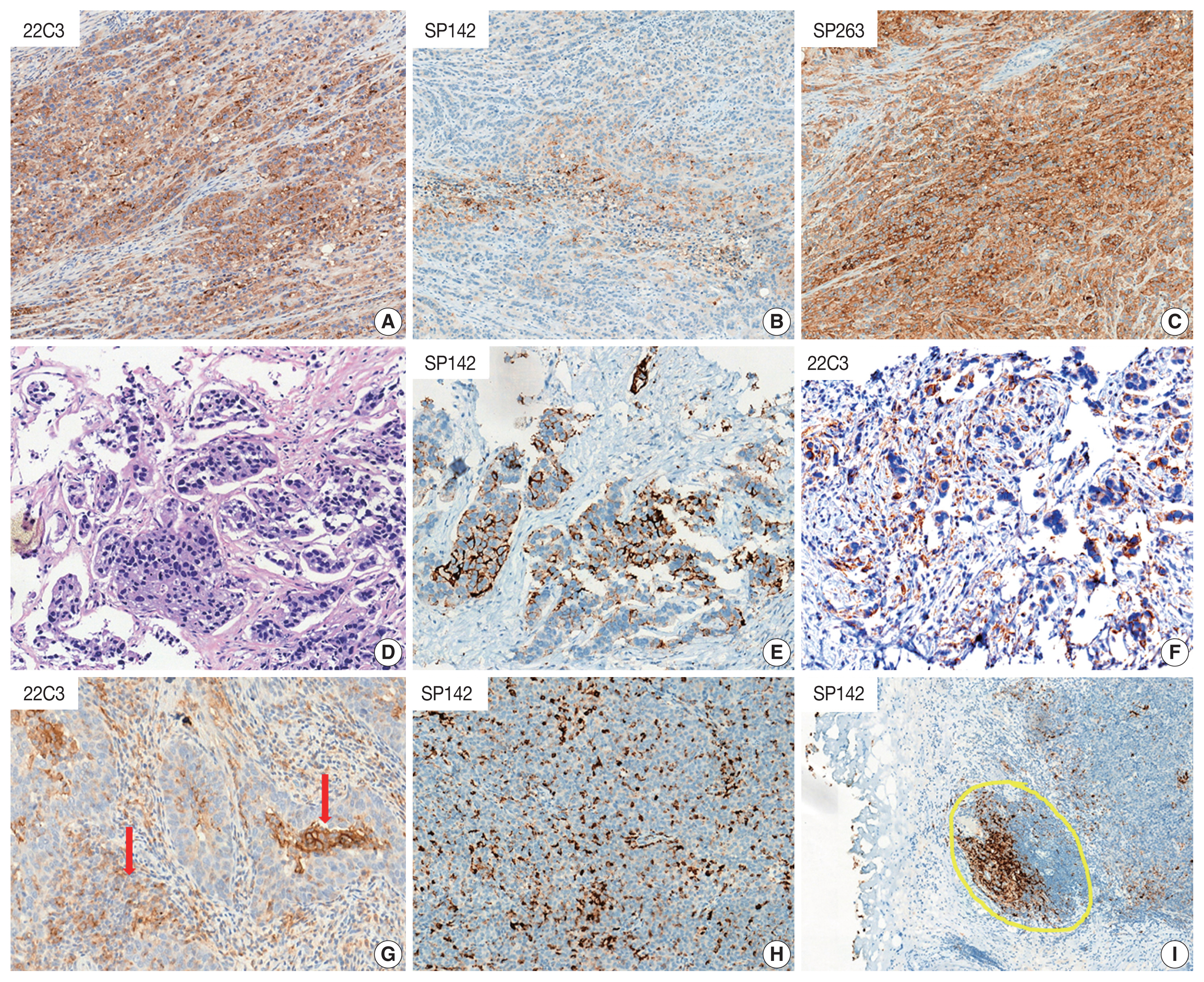
PDF - Programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) inhibition has revolutionized the treatment paradigm of urothelial carcinoma (UC). Several PD-L1 assays are conducted to formulate appropriate treatment decisions for PD-1/PD-L1 target therapy in UC. However, each assay has its own specific requirement of antibody clones, staining platforms, scoring algorithms, and cutoffs for the determination of PD-L1 status. These prove to be challenging constraints to pathology laboratories and pathologists. Thus, the present article comprehensively demonstrates the scoring algorithm used and differences observed in each assay (22C3, SP142, and SP263). Interestingly, the SP142 score algorithm considers only immune cells and not tumor cells (TCs). It remains controversial whether SP142 expressed only in TCs truly accounts for a negative PD-L1 case. Moreover, the scoring algorithm of each assay is complex and divergent, which can result in inter-observer heterogeneity. In this regard, the development of artificial intelligence for providing assistance to pathologists in obtaining more accurate and objective results has been actively researched. To facilitate efficiency of PD-L1 testing, several previous studies attempted to integrate and harmonize each assay in UC. The performance comparison of the various PD-L1 assays demonstrated in previous studies was encouraging, the exceptional concordance rate reported between 22C3 and SP263. Although these two assays may be used interchangeably, a clinically validated algorithm for each agent must be applied.
-
Citations
Citations to this article as recorded by- Comparison of tissue biomarkers between non-schistosoma and schistosoma-associated urothelial carcinoma
Nashwah Samir AlHariry, Enas A. El Saftawy, Basma Emad Aboulhoda, Ahmed H. Abozamel, Mansour A. Alghamdi, Amany E. Hamoud, Walaa Abd Elgawad Khalil Ghanam
Tissue and Cell.2024; 88: 102416. CrossRef - Aspectos prácticos sobre la determinación de PD-L1 en el tratamiento de carcinoma urotelial. Consenso del grupo de uropatología de la SEAP
Antonio López-Beltrán, Pilar González-Peramato, Julián Sanz-Ortega, Juan Daniel Prieto Cuadra, Isabel Trias, Rafael J. Luque Barona, María Eugenia Semidey, Pablo Maroto, Ferran Algaba
Revista Española de Patología.2023; 56(4): 261. CrossRef - Systemic treatment of advanced and metastatic urothelial cancer: The landscape in Australia
Howard Gurney, Timothy D. Clay, Niara Oliveira, Shirley Wong, Ben Tran, Carole Harris
Asia-Pacific Journal of Clinical Oncology.2023; 19(6): 585. CrossRef - PD-L1 Testing in Urothelial Carcinoma: Analysis of a Series of 1401 Cases Using Both the 22C3 and SP142 Assays
Harriet Evans, Brendan O’Sullivan, Frances Hughes, Kathryn Charles, Lee Robertson, Philippe Taniere, Salvador Diaz-Cano
Pathology and Oncology Research.2022;[Epub] CrossRef - Insights on recent innovations in bladder cancer immunotherapy
Mohamed A. Abd El‐Salam, Claire E.P. Smith, Chong‐Xian Pan
Cancer Cytopathology.2022; 130(9): 667. CrossRef - What Do We Have to Know about PD-L1 Expression in Prostate Cancer? A Systematic Literature Review. Part 1: Focus on Immunohistochemical Results with Discussion of Pre-Analytical and Interpretation Variables
Andrea Palicelli, Martina Bonacini, Stefania Croci, Cristina Magi-Galluzzi, Sofia Cañete-Portillo, Alcides Chaux, Alessandra Bisagni, Eleonora Zanetti, Dario De Biase, Beatrice Melli, Francesca Sanguedolce, Moira Ragazzi, Maria Paola Bonasoni, Alessandra
Cells.2021; 10(11): 3166. CrossRef
- Comparison of tissue biomarkers between non-schistosoma and schistosoma-associated urothelial carcinoma

 E-submission
E-submission

 First
First Prev
Prev



