Search
- Page Path
- HOME > Search
- KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay
- Sung Hak Lee, Arthur Minwoo Chung, Ahwon Lee, Woo Jin Oh, Yeong Jin Choi, Youn-Soo Lee, Eun Sun Jung
- J Pathol Transl Med. 2017;51(1):24-31. Published online December 25, 2016
- DOI: https://doi.org/10.4132/jptm.2016.10.03
- 12,776 View
- 170 Download
- 5 Web of Science
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Mutations in the KRAS gene have been identified in approximately 50% of colorectal cancers (CRCs). KRAS mutations are well established biomarkers in anti–epidermal growth factor receptor therapy. Therefore, assessment of KRAS mutations is needed in CRC patients to ensure appropriate treatment.
Methods
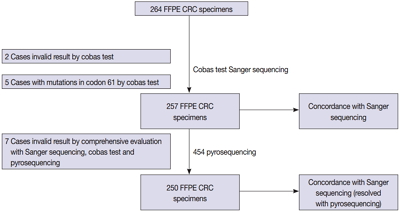
We compared the analytical performance of the cobas test to Sanger sequencing in 264 CRC cases. In addition, discordant specimens were evaluated by 454 pyrosequencing.
Results
KRAS mutations for codons 12/13 were detected in 43.2% of cases (114/264) by Sanger sequencing. Of 257 evaluable specimens for comparison, KRAS mutations were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% positive percent agreement (PPA) and 91.0% negative percent agreement (NPA) for codons 12/13. Results from the cobas test and Sanger sequencing were discordant for 20 cases (7.8%). Twenty discrepant cases were subsequently subjected to 454 pyrosequencing. After comprehensive analysis of the results from combined Sanger sequencing–454 pyrosequencing and the cobas test, PPA was 97.5% and NPA was 100%.
Conclusions
The cobas test is an accurate and sensitive test for detecting KRAS-activating mutations and has analytical power equivalent to Sanger sequencing. Prescreening using the cobas test with subsequent application of Sanger sequencing is the best strategy for routine detection of KRAS mutations in CRC. -
Citations
Citations to this article as recorded by- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
Journal of Pathology and Translational Medicine.2023; 57(4): 217. CrossRef - Assessment of KRAS and NRAS status in metastatic colorectal cancer: Experience of the National Institute of Oncology in Rabat Morocco
Chaimaa Mounjid, Hajar El Agouri, Youssef Mahdi, Abdelilah Laraqui, En-nacer Chtati, Soumaya Ech-charif, Mouna Khmou, Youssef Bakri, Amine Souadka, Basma El Khannoussi
Annals of Cancer Research and Therapy.2022; 30(2): 80. CrossRef - The current understanding on the impact of KRAS on colorectal cancer
Mingjing Meng, Keying Zhong, Ting Jiang, Zhongqiu Liu, Hiu Yee Kwan, Tao Su
Biomedicine & Pharmacotherapy.2021; 140: 111717. CrossRef - Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer
Barbora Vanova, Michal Kalman, Karin Jasek, Ivana Kasubova, Tatiana Burjanivova, Anna Farkasova, Peter Kruzliak, Dietrich Busselberg, Lukas Plank, Zora Lasabova
Clinical and Experimental Medicine.2019; 19(2): 219. CrossRef - CRISPR Technology for Breast Cancer: Diagnostics, Modeling, and Therapy
Rachel L. Mintz, Madeleine A. Gao, Kahmun Lo, Yeh‐Hsing Lao, Mingqiang Li, Kam W. Leong
Advanced Biosystems.2018;[Epub] CrossRef
- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
- Comparison of Direct Sequencing, PNA Clamping-Real Time Polymerase Chain Reaction, and Pyrosequencing Methods for the Detection of
EGFR Mutations in Non-small Cell Lung Carcinoma and the Correlation with Clinical Responses to EGFR Tyrosin - Hyun Ju Lee, Xianhua Xu, Hyojin Kim, Yan Jin, Pingli Sun, Ji Eun Kim, Jin-Haeng Chung
- Korean J Pathol. 2013;47(1):52-60. Published online February 25, 2013
- DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.1.52
- 13,048 View
- 93 Download
- 31 Crossref
-
 Abstract
Abstract
 PDF
PDF Background The aims of this study were to evaluate the abilities of direct sequencing (DS), peptide nucleic acid (PNA) clamping, and pyrosequencing methods to detect epidermal growth factor receptor (
EGFR ) mutations in formalin-fixed paraffin-embedded (FFPE) non-small cell lung carcinoma (NSCLC) samples and to correlateEGFR mutational status as determined by each method with the clinical response toEGFR tyrosine kinase inhibitors (TKIs).Methods Sixty-one NSCLC patients treated with EGFR TKIs were identified to investigate somatic mutations in the
EGFR gene (exons 18-21).Results Mutations in the
EGFR gene were detected in 38 of the 61 patients (62%) by DS, 35 (57%) by PNA clamping and 37 (61%) by pyrosequencing. A total of 44 mutations (72%) were found by at least one of the three methods, and the concordances among the results were relatively high (82-85%; kappa coefficient, 0.713 to 0.736). There were 15 discordant cases (25%) among the three different methods.Conclusions All three
EGFR mutation tests had good concordance rates (over 82%) for FFPE samples. These results suggest that if the DNA quality and enrichment of tumor cells are assured, then DS, PNA clamping, and pyrosequencing are appropriate methods for the detection ofEGFR mutations.-
Citations
Citations to this article as recorded by- Recent Trends of Lung Cancer in Korea
Jae Guk Lee, Ho Cheol Kim, Chang-Min Choi
Tuberculosis and Respiratory Diseases.2021; 84(2): 89. CrossRef - Predictive value of KRAS mutation and excision repair cross-complementing 1 (ERCC1) protein overexpression in patients with colorectal cancer administered FOLFOX regimen
Sun Min Park, Sung Bong Choi, Yoon Suk Lee, In Kyu Lee
Asian Journal of Surgery.2021; 44(5): 715. CrossRef - Recent advances in diagnostic technologies in lung cancer
Hye Jung Park, Sang Hoon Lee, Yoon Soo Chang
The Korean Journal of Internal Medicine.2020; 35(2): 257. CrossRef - Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p.L747P and p.L747S mutations
Sheng-Kai Liang, Jen-Chung Ko, James Chih-Hsin Yang, Jin-Yuan Shih
Lung Cancer.2019; 133: 103. CrossRef - Biomarker Testing for Patients With Advanced Non–Small Cell Lung Cancer: Real-World Issues and Tough Choices
Nathan A. Pennell, Maria E. Arcila, David R. Gandara, Howard West
American Society of Clinical Oncology Educational Book.2019; (39): 531. CrossRef - Evaluation of EGFR mutations in NSCLC with highly sensitive droplet digital PCR assays
Xi‑Wen Jiang, Wei Liu, Xiao‑Ya Zhu, Xiao‑Xie Xu
Molecular Medicine Reports.2019;[Epub] CrossRef - Peptide Nucleic Acid Clamping and Direct Sequencing in the Detection of Oncogenic Alterations in Lung Cancer: Systematic Review and Meta-Analysis
Jae-Uk Song, Jonghoo Lee
Yonsei Medical Journal.2018; 59(2): 211. CrossRef - Distribution of KRAS, DDR2, and TP53 gene mutations in lung cancer: An analysis of Iranian patients
Zahra Fathi, Seyed Ali Javad Mousavi, Raheleh Roudi, Farideh Ghazi, Sumitra Deb
PLOS ONE.2018; 13(7): e0200633. CrossRef - EGFR T790M mutation testing within the osimertinib AURA Phase I study
Simon Dearden, Helen Brown, Suzanne Jenkins, Kenneth S. Thress, Mireille Cantarini, Rebecca Cole, Malcolm Ranson, Pasi A. Jänne
Lung Cancer.2017; 109: 9. CrossRef - Molecular Testing of Lung Cancers
Hyo Sup Shim, Yoon-La Choi, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Mee Sook Roh, Tae-Jung Kim, Seung Yeon Ha, Jin-Haeng Chung, Se Jin Jang, Geon Kook Lee
Journal of Pathology and Translational Medicine.2017; 51(3): 242. CrossRef - Mutations of the Epidermal Growth Factor Receptor Gene in Triple-Negative Breast Cancer
Aeri Kim, Min Hye Jang, Soo Jung Lee, Young Kyung Bae
Journal of Breast Cancer.2017; 20(2): 150. CrossRef - Double primary lung adenocarcinoma diagnosed by epidermal growth factor receptor mutation status
Oh Jung Kwon, Min Hyeok Lee, Sung Ju Kang, Seul Gi Kim, In Beom Jeong, Ji Yun Jeong, Eun Jung Cha, Do Yeun Cho, Young Jin Kim, Ji Woong Son
Yeungnam University Journal of Medicine.2017; 34(2): 270. CrossRef - Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing
Mi-Young Park, Min Hee Jung, Eun Young Eo, Seokjoong Kim, Sang Hoon Lee, Yeon Joo Lee, Jong Sun Park, Young Jae Cho, Jin Haeng Chung, Cheol Hyeon Kim, Ho Il Yoon, Jae Ho Lee, Choon-Taek Lee
Oncotarget.2017; 8(22): 36331. CrossRef - Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study
Kyueng-Whan Min, Wan-Seop Kim, Se Jin Jang, Yoo Duk Choi, Sunhee Chang, Soon Hee Jung, Lucia Kim, Mee Sook Roh, Choong Sik Lee, Jung Weon Shim, Mi Jin Kim, Geon Kook Lee
QJM.2016; 109(3): 167. CrossRef - Epidermal Growth Factor Receptor Mutation and Anaplastic Lymphoma Kinase Gene Fusion: Detection in Malignant Pleural Effusion by RNA or PNA Analysis
Yi-Lin Chen, Chung-Ta Lee, Cheng-Chan Lu, Shu-Ching Yang, Wan-Li Chen, Yang-Cheng Lee, Chung-Hsien Yang, Shu-Ling Peng, Wu-Chou Su, Nan-Haw Chow, Chung-Liang Ho, Javier S Castresana
PLOS ONE.2016; 11(6): e0158125. CrossRef - IDH Mutation Analysis in Ewing Sarcoma Family Tumors
Ki Yong Na, Byeong-Joo Noh, Ji-Youn Sung, Youn Wha Kim, Eduardo Santini Araujo, Yong-Koo Park
Journal of Pathology and Translational Medicine.2015; 49(3): 257. CrossRef - Immunohistochemical demonstration of alteration of β-catenin during tumor metastasis by different mechanisms according to histology in lung cancer
XIANHUA XU, JI EUN KIM, PING-LI SUN, SEOL BONG YOO, HYOJIN KIM, YAN JIN, JIN-HAENG CHUNG
Experimental and Therapeutic Medicine.2015; 9(2): 311. CrossRef - Detection of EGFR-TK Domain–activating Mutations in NSCLC With Generic PCR-based Methods
Rajendra B. Shahi, Sylvia De Brakeleer, Jacques De Grève, Caroline Geers, Peter In’t Veld, Erik Teugels
Applied Immunohistochemistry & Molecular Morphology.2015; 23(3): 163. CrossRef - Frequent aerogenous spread with decreased E-cadherin expression of ROS1- rearranged lung cancer predicts poor disease-free survival
Yan Jin, Ping-Li Sun, Soo Young Park, Hyojin Kim, Eunhyang Park, Gilhyang Kim, Sukki Cho, Kwhanmien Kim, Choon-Taek Lee, Jin-Haeng Chung
Lung Cancer.2015; 89(3): 343. CrossRef - Membranous Insulin-like Growth Factor-1 Receptor (IGF1R) Expression Is Predictive of Poor Prognosis in Patients with Epidermal Growth Factor Receptor (EGFR)-Mutant Lung Adenocarcinoma
Eunhyang Park, Soo Young Park, Hyojin Kim, Ping-Li Sun, Yan Jin, Suk Ki Cho, Kwhanmien Kim, Choon-Taek Lee, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2015; 49(5): 382. CrossRef - Peptide Nucleic Acid Clamping Versus Direct Sequencing for the Detection of EGFR Gene Mutation in Patients with Non-small Cell Lung Cancer
Seong-Hoon Yoon, Yoo-Duk Choi, In-Jae Oh, Kyu-Sik Kim, Hayoung Choi, Jinsun Chang, Hong-Joon Shin, Cheol-Kyu Park, Young-Chul Kim
Cancer Research and Treatment.2015; 47(4): 661. CrossRef - Analysis of Mutations in Epidermal Growth Factor Receptor Gene in Korean Patients with Non-small Cell Lung Cancer: Summary of a Nationwide Survey
Sang Hwa Lee, Wan Seop Kim, Yoo Duk Choi, Jeong Wook Seo, Joung Ho Han, Mi Jin Kim, Lucia Kim, Geon Kook Lee, Chang Hun Lee, Mee Hye Oh, Gou Young Kim, Sun Hee Sung, Kyo Young Lee, Sun Hee Chang, Mee Sook Rho, Han Kyeom Kim, Soon Hee Jung, Se Jin Jang
Journal of Pathology and Translational Medicine.2015; 49(6): 481. CrossRef - Novel EGFR mutation-specific antibodies for lung adenocarcinoma: Highly specific but not sensitive detection of an E746_A750 deletion in exon 19 and an L858R mutation in exon 21 by immunohistochemistry
An Na Seo, Tae-In Park, Yan Jin, Ping-Li Sun, Hyojin Kim, Hyun Chang, Jin-Haeng Chung
Lung Cancer.2014; 83(3): 316. CrossRef - Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non‐small‐cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage
Tae‐Jung Kim, Chan Kwon Park, Chang Dong Yeo, Kihoon Park, Chin Kook Rhee, Jusang Kim, Seung Joon Kim, Sang Haak Lee, Kyo‐Young Lee, Hyoung‐Kyu Yoon
Journal of Surgical Oncology.2014; 110(3): 245. CrossRef - Epidermal growth factor receptor mutations and anaplastic lymphoma kinase rearrangements in lung cancer with nodular ground-glass opacity
Sung-Jun Ko, Yeon Joo Lee, Jong Sun Park, Young-Jae Cho, Ho Il Yoon, Jin-Haeng Chung, Tae Jung Kim, Kyung Won Lee, Kwhanmien Kim, Sanghoon Jheon, Hyojin Kim, Jae Ho Lee, Choon-Taek Lee
BMC Cancer.2014;[Epub] CrossRef - Cytoplasmic YAP Expression is Associated with Prolonged Survival in Patients with Lung Adenocarcinomas and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment
Ping-Li Sun, Ji Eun Kim, Seol Bong Yoo, Hyojin Kim, Yan Jin, Sanghoon Jheon, Kwhanmien Kim, Choon Taek Lee, Jin-Haeng Chung
Annals of Surgical Oncology.2014; 21(S4): 610. CrossRef - Sensitive methods for detection of the S768R substitution in exon 18 of the DDR2 gene in patients with central nervous system metastases of non-small cell lung cancer
Marcin Nicoś, Tomasz Powrózek, Paweł Krawczyk, Bożena Jarosz, Beata Pająk, Marek Sawicki, Krzysztof Kucharczyk, Tomasz Trojanowski, Janusz Milanowski
Medical Oncology.2014;[Epub] CrossRef - Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas
A N Seo, J M Yang, H Kim, S Jheon, K Kim, C T Lee, Y Jin, S Yun, J-H Chung, J H Paik
British Journal of Cancer.2014; 110(11): 2688. CrossRef - KRASMutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing
Boram Lee, Boin Lee, Gangmin Han, Mi Jung Kwon, Joungho Han, Yoon-La Choi
Korean Journal of Pathology.2014; 48(2): 100. CrossRef - Guideline Recommendations forEGFRMutation Testing in Lung Cancer: Proposal of the Korean Cardiopulmonary Pathology Study Group
Hyo Sup Shim, Jin-Haeng Chung, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Geon Kook Lee, Soon-Hee Jung, Se Jin Jang
Korean Journal of Pathology.2013; 47(2): 100. CrossRef - Immunohistochemical Classification of Primary and Secondary Glioblastomas
Kyu Sang Lee, Gheeyoung Choe, Kyung Han Nam, An Na Seo, Sumi Yun, Kyung Ju Kim, Hwa Jin Cho, Sung Hye Park
Korean Journal of Pathology.2013; 47(6): 541. CrossRef
- Recent Trends of Lung Cancer in Korea
- MGMT Gene Promoter Methylation Analysis by Pyrosequencing of Brain Tumour.
- Young Zoon Kim, Young Jin Song, Ki Uk Kim, Dae Cheol Kim
- Korean J Pathol. 2011;45(5):455-462.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.5.455
- 4,831 View
- 16 Download
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
The aim of this study was to determine whether pyrosequencing (PSQ) might be useful to achieve O6-methyl guanine methyltransferase (MGMT) promoter methylation using 1- to 13-year-old archival tissues as a clinical biomarker in routine practice.
METHODS
The study included 141 formalin-fixed paraffin-embedded (FFPE) glial tumors from the archives of the Pathology Department from 1997-2010.
RESULTS
The average percentage of methylation (MP) of the 141 cases was 14.0+/-16.8%, and methylated cases were 32.3+/-14.9%. The average MP of each year did not show a linear increasing or decreasing pattern according to the age of the FFPE block (p=0.771). The average MP of methylated glioblastomas was 35.8+/-14.7%, 31.8+/-15.5% for anaplastic astrocytomas, and 22.4+/-15.1% for astrocytoma. A tendency was observed toward an increasing pattern of average MP with World Health Organization (WHO) grade (p=0.063) in astrocytic tumors. A correlation was observed between average MP and WHO grade (p=0.038) and a bimodal distribution was observed between the methylated and unmethylated cases, using a 9% cut-off value (p<0.001).
CONCLUSIONS
The results showed that a quantitative approach for MGMT promoter methylation yielded a 100% success rate for FFPE tissues from archives. PSQ can be used in a retrospective trial, but the cut-off value and calculation method should be further validated. -
Citations
Citations to this article as recorded by- CDKN2A Homozygous Deletion Is a Stronger Predictor of Outcome than IDH1/2-Mutation in CNS WHO Grade 4 Gliomas
Sang Hyuk Lee, Tae Gyu Kim, Kyeong Hwa Ryu, Seok Hyun Kim, Young Zoon Kim
Biomedicines.2024; 12(10): 2256. CrossRef - Immunohistochemical Classification of Primary and Secondary Glioblastomas
Kyu Sang Lee, Gheeyoung Choe, Kyung Han Nam, An Na Seo, Sumi Yun, Kyung Ju Kim, Hwa Jin Cho, Sung Hye Park
Korean Journal of Pathology.2013; 47(6): 541. CrossRef
- CDKN2A Homozygous Deletion Is a Stronger Predictor of Outcome than IDH1/2-Mutation in CNS WHO Grade 4 Gliomas
- A Consideration of MGMT Gene Promotor Methylation Analysis for Glioblastoma Using Methylation-Specific Polymerase Chain Reaction and Pyrosequencing.
- Sang Hwa Lee, Tae Sook Hwang, Young Cho Koh, Wook Youn Kim, Hye Seung Han, Wan Seop Kim, Young Sin Ko, So Dug Lim
- Korean J Pathol. 2011;45(1):21-29.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.1.21
- 5,219 View
- 52 Download
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
O6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation is currently the most promising predictive marker for the outcome and benefit from temozolomide treatment in patients with glioblastoma, but there is no consensus on the analysis method for assessing the methylation status in the molecular diagnostic field. The objective of this study was to evaluate methylation-specific polymerase chain reaction (MSP) and pyrosequencing methods for assessing MGMT gene promotor methylation of glioblastoma as well as assessing the MGMT protein expression by immunohistochemistry.
METHODS
Twenty-seven cases of glioblastoma from the archives at the Department of Pathology Konkuk University Hospital were selected. MGMT promoter methylation was evaluated by MSP and the pyrosequencing methods. The MGMT expression was also measured at the protein level by immunohistochemistry.
RESULTS
Overall, MGMT hypermethylation was observed in 44.4% (12/27 cases) of the case of glioblastoma using either MSP or pyrosequencing. The concordant rate was 70.3% (19/27 cases) between MSP and pyrosequencing for MGMT methylation. There was no correlation between MGMT methylation and the protein expression. No significant differences in progression free survival and overall survival were seen between the methylated group and the unmethylated group by using either MSP or pyrosequencing. The status of the MGMT protein expression was correlated with progression free survival (p=0.026).
CONCLUSIONS
In this study the concordance rate between MSP and the pyrosequencing methods for assessing MGMT gene promotor methylation was relatively low for the cases of glioblastoma. This suggests that more reliable techniques for routine MGMT methylation study of glioblastoma remain to be developed because of quality control and assurance issues. -
Citations
Citations to this article as recorded by- Prognostic Role of Methylation Status of theMGMTPromoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients
Dae Cheol Kim, Ki Uk Kim, Young Zoon Kim
Journal of Korean Neurosurgical Society.2016; 59(1): 26. CrossRef - Distinct genetic alterations in pediatric glioblastomas
Sun-ju Byeon, Jae Kyung Myung, Se Hoon Kim, Seung-Ki Kim, Ji Hoon Phi, Sung-Hye Park
Child's Nervous System.2012; 28(7): 1025. CrossRef - MGMTGene Promoter Methylation Analysis by Pyrosequencing of Brain Tumour
Young Zoon Kim, Young Jin Song, Ki Uk Kim, Dae Cheol Kim
The Korean Journal of Pathology.2011; 45(5): 455. CrossRef
- Prognostic Role of Methylation Status of theMGMTPromoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients

 E-submission
E-submission

 First
First Prev
Prev



