Search
- Page Path
- HOME > Search
Original Articles
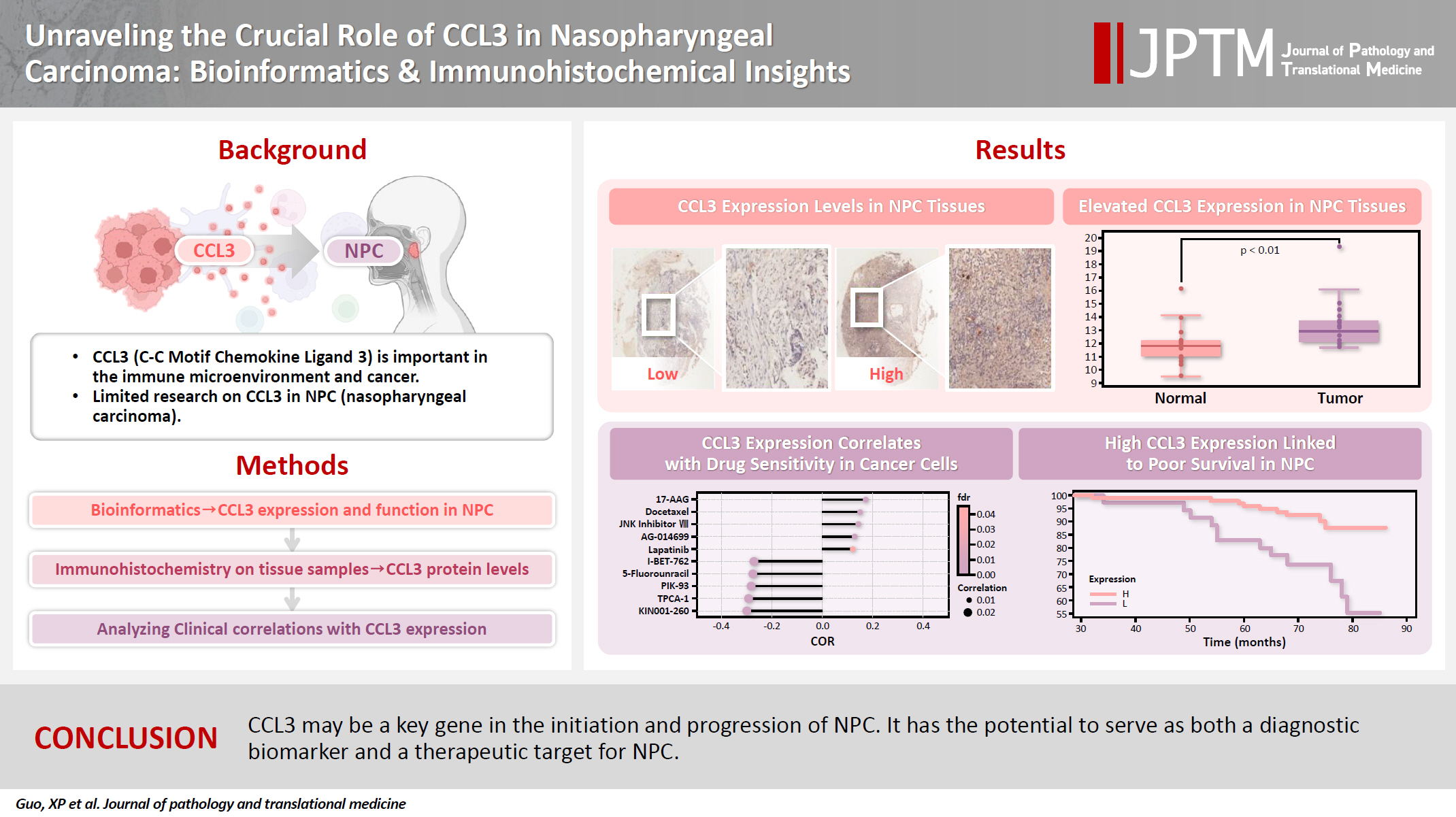
- Unraveling the crucial role of CCL3 in nasopharyngeal carcinoma: bioinformatics and immunohistochemical insights
- Xiaopeng Guo, Zhen Sun, Ya Liang, Aoshuang Chang, Junjun Ling, Houyu Zhao, Xianlu Zhuo
- J Pathol Transl Med. 2025;59(5):281-290. Published online September 8, 2025
- DOI: https://doi.org/10.4132/jptm.2025.05.23
- 1,540 View
- 138 Download
-
 Abstract
Abstract
 PDF
PDF - Background
C-C motif chemokine ligand 3 (CCL3) is a crucial chemokine that plays a fundamental role in the immune microenvironment and is closely linked to the development of various cancers. Despite its importance, there is limited research regarding the expression and function of CCL3 in nasopharyngeal carcinoma (NPC). Therefore, this study seeks to examine the expression of CCL3 and assess its clinical significance in NPC using bioinformatics analysis and experiments. Methods: The bioinformatics approach was employed to assess the expression and function of CCL3 in NPC. Subsequently, protein expression of CCL3 was detected in an NPC cohort using immunohistochemistry based on a tissue microarray. The relationship between CCL3 expression and clinical features was then investigated. Results: A total of 20 CCL3-related genes and 14 possible target genes were identified through bioinformatics analysis, many of which play crucial roles in pathways such as chemokine signaling pathway and transcriptional misregulation in cancer signaling pathways. CCL3 was found to be associated with drug resistance and various immune cell infiltrations. In NPC, CCL3 expression was significantly higher than normal controls, and high expression of CCL3 correlated with cervical lymph node metastasis, tumor recurrence, advanced clinical stage, and poor prognosis. Conclusions: CCL3 may be a key gene in the initiation and progression of NPC. It has the potential to serve as both a diagnostic biomarker and a therapeutic target for NPC.
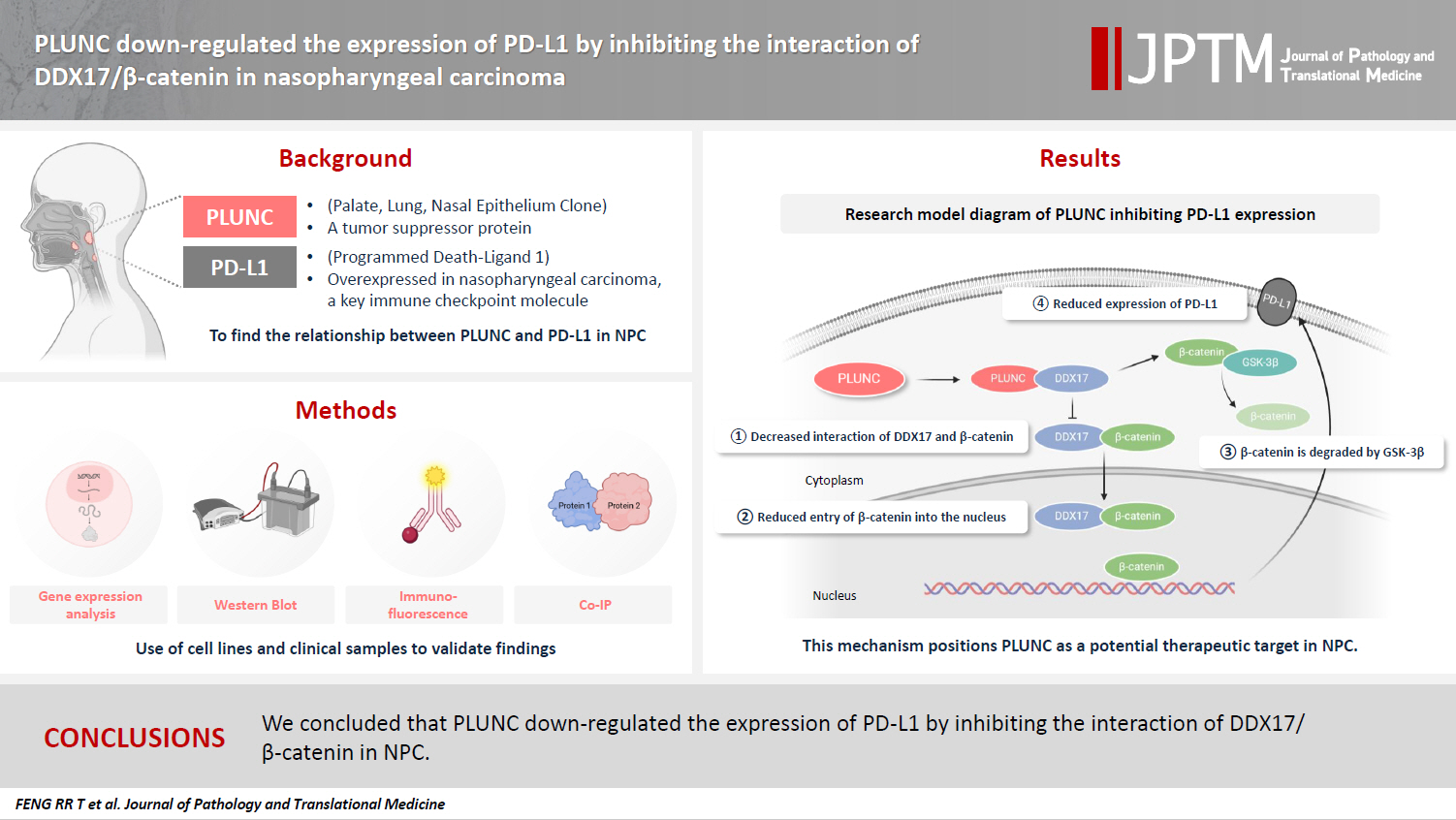
- PLUNC downregulates the expression of PD-L1 by inhibiting the interaction of DDX17/β-catenin in nasopharyngeal carcinoma
- Ranran Feng, Yilin Guo, Meilin Chen, Ziying Tian, Yijun Liu, Su Jiang, Jieyu Zhou, Qingluan Liu, Xiayu Li, Wei Xiong, Lei Shi, Songqing Fan, Guiyuan Li, Wenling Zhang
- J Pathol Transl Med. 2025;59(1):68-83. Published online January 15, 2025
- DOI: https://doi.org/10.4132/jptm.2024.11.27
- 3,449 View
- 137 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Nasopharyngeal carcinoma (NPC) is characterized by high programmed death-ligand 1 (PD-L1) expression and abundant infiltration of non-malignant lymphocytes, which renders patients potentially suitable candidates for immune checkpoint blockade therapies. Palate, lung, and nasal epithelium clone (PLUNC) inhibit the growth of NPC cells and enhance cellular apoptosis and differentiation. Currently, the relationship between PLUNC (as a tumor-suppressor) and PD-L1 in NPC is unclear.
Methods
We collected clinical samples of NPC to verify the relationship between PLUNC and PD-L1. PLUNC plasmid was transfected into NPC cells, and the variation of PD-L1 was verified by western blot and immunofluorescence. In NPC cells, we verified the relationship of PD-L1, activating transcription factor 3 (ATF3), and β-catenin by western blot and immunofluorescence. Later, we further verified that PLUNC regulates PD-L1 through β-catenin. Finally, the effect of PLUNC on β-catenin was verified by co-immunoprecipitation (Co-IP).
Results
We found that PLUNC expression was lower in NPC tissues than in paracancer tissues. PD-L1 expression was opposite to that of PLUNC. Western blot and immunofluorescence showed that β-catenin could upregulate ATF3 and PD-L1, while PLUNC could downregulate ATF3/PD-L1 by inhibiting the expression of β-catenin. PLUNC inhibits the entry of β-catenin into the nucleus. Co-IP experiments demonstrated that PLUNC inhibited the interaction of DEAD-box helicase 17 (DDX17) and β-catenin.
Conclusions
PLUNC downregulates the expression of PD-L1 by inhibiting the interaction of DDX17/β-catenin in NPC. -
Citations
Citations to this article as recorded by- The Potential Role of SP-G and PLUNC in Tumor Pathogenesis and Wound Healing in the Human Larynx
Aurelius Scheer, Lars Bräuer, Markus Eckstein, Heinrich Iro, Friedrich Paulsen, Fabian Garreis, Martin Schicht, Antoniu-Oreste Gostian
Biomedicines.2025; 13(5): 1240. CrossRef - Role of DEAD/DEAH-box helicases in immunity, infection and cancers
Rex Devasahayam Arokia Balaya, Saptami Kanekar, Shreya Kumar, Richard K. Kandasamy
Cell Communication and Signaling.2025;[Epub] CrossRef - CHIP modulates Wnt/β-catenin signalling in colorectal cancer through proteasomal degradation of DDX17
Sunny Kumar, Sayani Ghosh, Malini Basu, Mrinal K. Ghosh
Biochimica et Biophysica Acta (BBA) - Molecular Cell Research.2025; 1872(8): 120049. CrossRef
- The Potential Role of SP-G and PLUNC in Tumor Pathogenesis and Wound Healing in the Human Larynx
- Expression of Epstein-Barr Virus Gene Products, Bcl-2 and p53 Proteins in Nasopharyngeal Carcinomas.
- Sun Hee Yoon, Kang Suek Suh, Chang Hun Lee
- Korean J Pathol. 1997;31(8):723-734.
- 1,972 View
- 16 Download
-
 Abstract
Abstract
 PDF
PDF - The authors studied EBV genome expression in 40 conventionally processed samples of nasopharyngeal carcinomas (NPC), using in situ hybridization for EBERs and immunohistochemistry for LMP, Bcl-2 and p53 proteins. The NPCs consisted of 6 keratinizing squamous cell carcinomas (KSCs), 13 nonkeratinizing carcinomas (NKCs) and 21 undifferentiated carcinomas (UCs). The results were summarized as follows: 1) EBERs were expressed in 80.0% of all the NPCs (32/40). As for the subtypes, they were detected in 92.3% of NKCs (12/13), in 90.5% of the UCs (19/21), and in 16.7% of the KSCs (1/6). In positive cases, the nuclei of tumor cells displayed uniformly strong staining. 2) LMP was expressed in 10.0% of all the NPCs (4/40), all of which were UC. The LMP expression in the UCs was not correlated to the expression of EBERs, Bcl-2 and p53 proteins. 3) Bcl-2 protein was detected in 85.0% of all the NPCs (34/40). As for the subtypes, they were detected in 92.3% of the NKCs (12/13), in 90.5% of the UCs (19/21), and in 50.0% of the KSCs (3/6). 4) p53 protein was detected in 75.0% of all the NPCs (30/40). As for the subtypes, they were detected in 81.0% of the UCs (17/21), in 69.2% of the NKCs (9/13), and in 66.7% of the KSCs (4/6). 5) In the NPCs the expression of EBER showed a significantly positive correlation with that of p53 or Bcl-2 protein. The above results indicate that the association of EBV with NPC is chiefly with poorly differentiated and undifferentiated carcinomas. Additionally, carcinomas commonly display widespread, strong immunoreactivity of Bcl-2 and p53 proteins over tumor cells. In conclusion, these observations indicate that the EBV-association in NPC appears to contribute to the overexpression of tumor-related genes during carcinogenesis.
- Epstein-Barr Virus and p53 in Laryngeal and Nasopharyngeal Carcinomas.
- Eun Sook Nam, Duck Hwan Kim, Hyung Sik Shin, Young Euy Park, Young Sik Kim, Insun Kim
- Korean J Pathol. 1998;32(8):551-562.
- 2,137 View
- 10 Download
-
 Abstract
Abstract
- To investigate the correlation between EBV infection and p53 overexpression in laryngeal carcinomas (LC) and nasopharyngeal carcinomas (NPC) in Korea, we analyzed 37 laryngeal squamous cell carcinomas and 33 nasopharyngeal (11 squamous cell and 22 undifferentiated) carcinomas. We used the immunohistochemistry and polymerase chain reaction-single stranded conformational polymorphism (PCR-SSCP) for p53 overexpression and p53 gene mutation, respectively, and EBER-1 in situ hybridization and PCR using primer for EBNA-1 and EBNA-2 type 1 and 2 for prevalence and the subtype of EBV. The results were as follows; 1) The p53 expression was found in 43.2% of squamous cell LCs, in 54.6% of squamous cell NPCs and in 22.7% of undifferentiated NPCs. The p53 gene mutation was detected in 6 of 23 squamous cell LCs and 3 of 14 undifferentiated NPCs. 2) EBV was detected more frequently in undifferentiated NPCs (95.5%) than in squamous cell NPCs (63.6%) and squamous cell LCs (37.0%). Only type 1 was found in squamous cell LCs and NPCs, whereas both type 1 and type 2 were detected in undifferentiated NPCs. 3) There was no difference according to EBV infection (EBV+ ; 7 cases, EBV- ; 7 cases) in the cases with p53 protein overexpression but mutaion. From the above results, it can be concluded that squamous cell LCs and NPCs are associated with both p53 and EBV, whereas undifferentiated NPCs are more closely associated with EBV than p53. In Korea, both type 1 and 2 are detected in undifferentiated NPCs. Also, our result suggests that EBV infection does not seem to contribute to p53 overexpression. The interrelationship between EBV infection and p53 remains to be further defined.

 E-submission
E-submission

 First
First Prev
Prev



