Search
- Page Path
- HOME > Search
Original Articles
- Categorizing high-grade serous ovarian carcinoma into clinically relevant subgroups using deep learning–based histomic clusters
- Byungsoo Ahn, Eunhyang Park
- J Pathol Transl Med. 2025;59(2):91-104. Published online February 18, 2025
- DOI: https://doi.org/10.4132/jptm.2024.10.23
- 5,086 View
- 252 Download
- 1 Web of Science
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
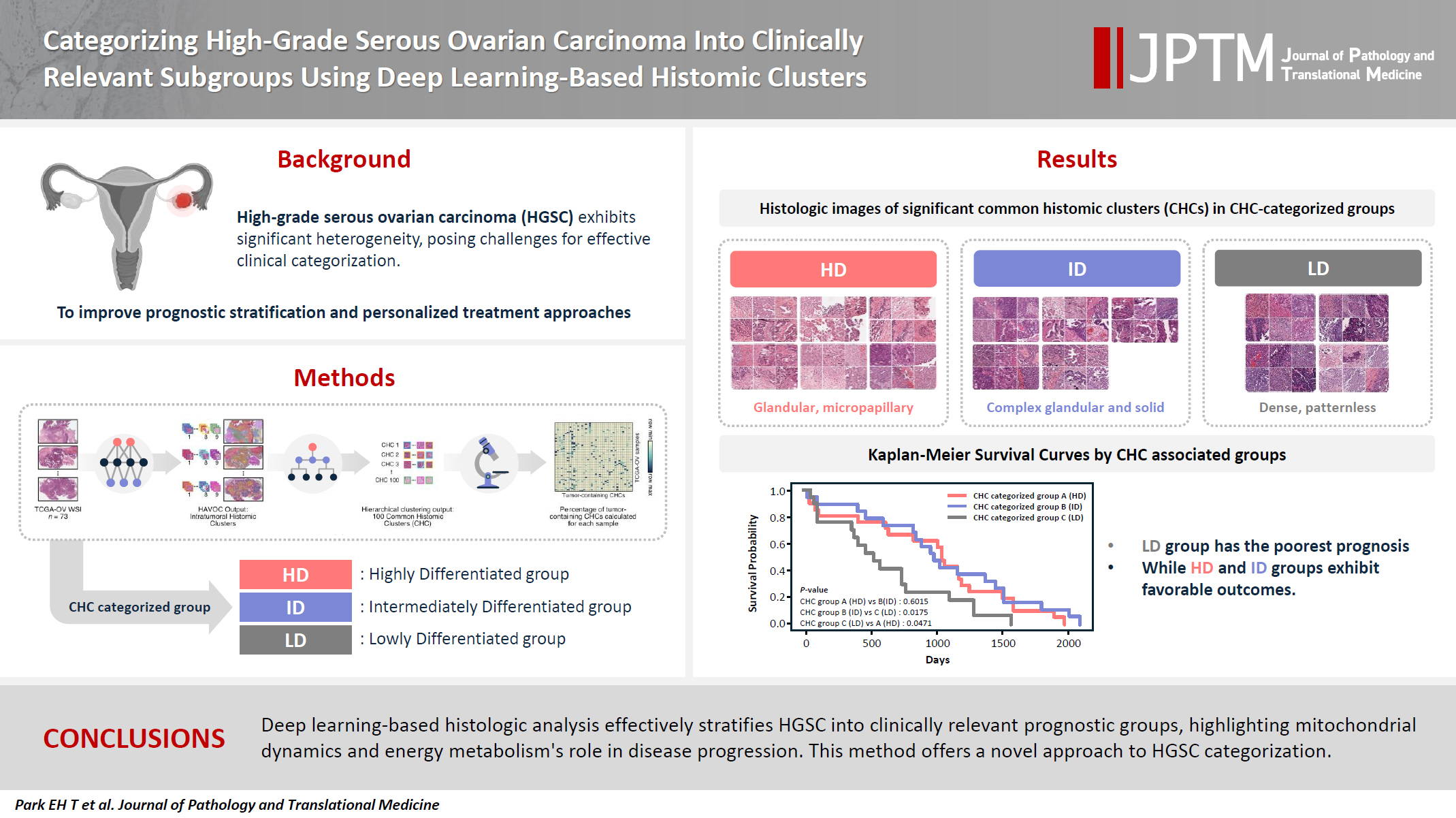
High-grade serous ovarian carcinoma (HGSC) exhibits significant heterogeneity, posing challenges for effective clinical categorization. Understanding the histomorphological diversity within HGSC could lead to improved prognostic stratification and personalized treatment approaches. Methods: We applied the Histomic Atlases of Variation Of Cancers model to whole slide images from The Cancer Genome Atlas dataset for ovarian cancer. Histologically distinct tumor clones were grouped into common histomic clusters. Principal component analysis and K-means clustering classified HGSC samples into three groups: highly differentiated (HD), intermediately differentiated (ID), and lowly differentiated (LD). Results: HD tumors showed diverse patterns, lower densities, and stronger eosin staining. ID tumors had intermediate densities and balanced staining, while LD tumors were dense, patternless, and strongly hematoxylin-stained. RNA sequencing revealed distinct patterns in mitochondrial oxidative phosphorylation and energy metabolism, with upregulation in the HD, downregulation in the LD, and the ID positioned in between. Survival analysis showed significantly lower overall survival for the LD compared to the HD and ID, underscoring the critical role of mitochondrial dynamics and energy metabolism in HGSC progression. Conclusions: Deep learning-based histologic analysis effectively stratifies HGSC into clinically relevant prognostic groups, highlighting the role of mitochondrial dynamics and energy metabolism in disease progression. This method offers a novel approach to HGSC categorization. -
Citations
Citations to this article as recorded by- Learning Disabilities in the 21st Century: Integrating Neuroscience, Education, and Technology for Better Outcomes
Syed Mohammed Basheeruddin Asdaq, Ahmad H. Alhowail, Syed Imam Rabbani, Naira Nayeem, Syed Mohammed Emaduddin Asdaq, Faiqa Nausheen
SAGE Open.2025;[Epub] CrossRef
- Learning Disabilities in the 21st Century: Integrating Neuroscience, Education, and Technology for Better Outcomes
- Senescent tumor cells in colorectal cancer are characterized by elevated enzymatic activity of complexes 1 and 2 in oxidative phosphorylation
- Jun Sang Shin, Tae-Gyu Kim, Young Hwa Kim, So Yeong Eom, So Hyun Park, Dong Hyun Lee, Tae Jun Park, Soon Sang Park, Jang-Hee Kim
- J Pathol Transl Med. 2023;57(6):305-314. Published online November 7, 2023
- DOI: https://doi.org/10.4132/jptm.2023.10.09
- 6,961 View
- 315 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Cellular senescence is defined as an irreversible cell cycle arrest caused by various internal and external insults. While the metabolic dysfunction of senescent cells in normal tissue is relatively well-established, there is a lack of information regarding the metabolic features of senescent tumor cells.
Methods
Publicly available single-cell RNA-sequencing data from the GSE166555 and GSE178341 datasets were utilized to investigate the metabolic features of senescent tumor cells. To validate the single-cell RNA-sequencing data, we performed senescence-associated β-galactosidase (SA-β-Gal) staining to identify senescent tumor cells in fresh frozen colorectal cancer tissue. We also evaluated nicotinamide adenine dinucleotide dehydrogenase–tetrazolium reductase (NADH-TR) and succinate dehydrogenase (SDH) activity using enzyme histochemical methods and compared the staining with SA-β-Gal staining. MTT assay was performed to reveal the complex 1 activity of the respiratory chain in in-vitro senescence model.
Results
Single-cell RNA-sequencing data revealed an upregulation in the activity of complexes 1 and 2 in oxidative phosphorylation, despite overall mitochondrial dysfunction in senescent tumor cells. Both SA-β-Gal and enzyme histochemical staining using fresh frozen colorectal cancer tissues indicated a high correlation between SA-β-Gal positivity and NADH-TR/SDH staining positivity. MTT assay showed that senescent colorectal cancer cells exhibit higher absorbance in 600 nm wavelength.
Conclusions
Senescent tumor cells exhibit distinct metabolic features, characterized by upregulation of complexes 1 and 2 in the oxidative phosphorylation pathway. NADH-TR and SDH staining represent efficient methods for detecting senescent tumor cells in colorectal cancer. -
Citations
Citations to this article as recorded by- Senescence, Aging and Disease Throughout the Gastrointestinal System
Sofia Ferreira-Gonzalez, Tomonori Matsumoto, Eiji Hara, Stuart J. Forbes
Gastroenterology.2025; 169(7): 1357. CrossRef - Cellular Aging and Senescence in Cancer: A Holistic Review of Cellular Fate Determinants
Muhammad Tufail, Yu-Qi Huang, Jia-Ju Hu, Jie Liang, Cai-Yun He, Wen-Dong Wan, Can-Hua Jiang, Hong Wu, Ning Li
Aging and disease.2024;[Epub] CrossRef - Real-time assessment of relative mitochondrial ATP synthesis response against inhibiting and stimulating substrates (MitoRAISE)
Eun Sol Chang, Kyoung Song, Ji-Young Song, Minjung Sung, Mi-Sook Lee, Jung Han Oh, Ji-Yeon Kim, Yeon Hee Park, Kyungsoo Jung, Yoon-La Choi
Cancer & Metabolism.2024;[Epub] CrossRef
- Senescence, Aging and Disease Throughout the Gastrointestinal System
- The Effect of Common Bile Duct Ligation on Liver Morphology and Coper Metabolism in Rat.
- Kyoung Sook Kim, Chanil Park, Jang Whan Cho, In Joon Choi, Yoo Bock Lee
- Korean J Pathol. 1990;24(4):402-411.
- 2,383 View
- 26 Download
-
 Abstract
Abstract
 PDF
PDF - To clarity the effect of biliary obliteration on copper metabolism of rat liver and on the hepatic morphology, 0.5% cuppuric sulfate was administered intraperitoneally for 42 days following ligation of the common bile duct (CBD) of Sprague-Dawley rats. The blood copper concentration, the hepatic copper content and the accumulation patterns of copper and copper binding protein in the liver were examined and compared with those of the simple CBD ligation group and the simple copper over loaded group. CBD ligation induced marked proliferation of bile ductular structures which, after expanding the portal tracts, invaded and divided the hepatic lobules. There was, however, no excess fibosis beyond what needed to support the new ductules. The blood copper concentration and the hepatic copper content were increased by copper overload with or without CBD ligation, particularly incases with CBD ligation. Liver cell necrosis did not occur by the overloaded copper alone in rats. The hepatic copper and copper binding protein were accumulated at periportal liver cells in the group of coppe overload after CBD ligatio, whereas they began to appear at perivenular hepatocytes in the simple copper overloaded group. In conclusion, it is suggested that CBD ligation does not induce excess fibrosis or liver cirrhosis in rat as far as during our experimental period, but affect significantly on copper metabolism by intrahepatic redistribution of the copper and the copper binding proteins.

 E-submission
E-submission


 First
First Prev
Prev



