Effects of Fixation and Storage of Human Tissue Samples on Nucleic Acid Preservation
Article information
Abstract
Background
Because of recent advances in the molecular diagnosis of cancer patients, tissue quality has become more important in daily practice.
Methods
To evaluate the effects of fixative, duration of fixation, decalcification, and storage periods on nucleic acid integrity, DNA and RNA were extracted from gastrointestinal cancer tissue. The yield and purity were analyzed, and polymerase chain reaction (PCR) for glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 60 bp), β-actin (148 bp), and human growth hormone (hGH; 434 bp) and real-time reverse transcription-PCR for β-actin (97 bp) were performed.
Results
All formalin-fixed paraffin-embedded (FFPE) and methacarn-fixed paraffin-embedded (MFPE) samples tested positive for GAPDH and β-actin by PCR. hGH was successfully detected in all MFPE samples, but in only 46.7% of the FFPE samples. Prolonged formalin fixation resulted in fewer GAPDH and β-actin PCR products, and amplification of hGH was not successful. The PCR and reverse transcription-PCR results were significantly affected by the duration of decalcification. The yield, purity, and integrity of mRNA progressively decreased with increased storage periods of paraffin blocks.
Conclusions
Fixation and storage should therefore be standardized in order to improve the quality of molecular pathologic diagnosis.
The use of molecular pathologic diagnosis for managing cancer patients has rapidly increased. The quality of the tissue analyzed, which varies according to tissue fixation and preservation, is thus important in pathologic laboratories. Genetic alterations and expression profiling of mRNA or protein are essential in diagnosing cancer and guiding treatment.1,2 In colorectal cancer patients, molecular examination of microsatellite instability is required to screen for hereditary nonpolyposis colorectal cancer and grade sporadic colorectal cancer.3,4 In addition, mutational analysis of KRAS, BRAF, and PIK3CA by sequencing or real-time polymerase chain reaction (PCR) is performed daily to predict the response to cetuximab therapy.5 The accuracy and reproducibility of this molecular diagnosis depend on the quantity and quality of nucleic acids obtained from cancer tissue specimens.6
Because there is no standard tissue fixation method, various methods are employed. Although freezing is considered one of the best methods of conserving nucleic acids and proteins, archival formalin-fixed paraffin-embedded (FFPE) tissue is widely used in clinical pathologic laboratories. However, the effects of formalin fixation on nucleic acids are known to be significant.7 Studies of chemical reactions between formalin and nucleic acids have demonstrated that the formalin initiates DNA denaturation in the AT-rich regions of double-stranded DNA and creates sites for chemical interaction. Furthermore, RNA may be degraded or chemically modified during fixation, thereby resulting in poor yields.7,8 Several alternative fixatives have therefore been developed and tested. Of these, methacarn has proven to be an excellent fixative for preserving nucleic acids of human samples.9 Damaged nucleic acid negatively affects analysis and diagnosis. In addition to the type of fixative, several other factors including the duration of fixation, decalcification, and the storage conditions (i.e., time, temperature, and humidity) of the paraffin blocks affect the quantity and quality of nucleic acids.10-13
In this study, we compared the efficacy of formalin and methacarn fixation in preserving nucleic acids in tissue samples. We also evaluated the effects of duration of fixation, decalcification, and storage time on the integrity of the nucleic acids.
MATERIALS AND METHODS
Samples and preparations
We selected 30 FFPE-fixed tissue samples from 2003 to 2011 and 15 methacarn-fixed paraffin-embedded (MFPE) tissue samples from 2009 to 2011 from the Department of Pathology at Seoul National University Bundang Hospital. All samples were from surgical resections of gastric or colorectal cancer. We used 10% neutral buffered formaldehyde (formalin; Samchun Chemicals, Pyeongtaek, Korea). The methacarn solution consisted of 60% methanol (Duksan Pure Chemicals, Ansan, Korea), 30% chloroform (Duksan Pure Chemicals), and 10% acetic acid (Duksan Pure Chemicals).
To investigate the effects of decalcification on the quantity and quality of nucleic acids, five samples of gastrointestinal cancer tissue were treated with decalcification solution for 0, 10, 30, 60, 120, and 180 minutes. To evaluate the effects of duration of fixation, we fixed the tissue in formalin for 3, 7, 30, 90, and 180 days at room temperature. In order to compare the influence of formalin, we fixed the tissue in formalin for one day and then stored them in 70% ethanol at 4℃ for 2, 6, 29, 89, and 179 days.
DNA and RNA extraction
Six 8-µm-thick FFPE or MFPE tissue sections were used for DNA extraction, and 11 FFPE or MFPE tissue sections were used for RNA. We microscopically dissected a 1×1 cm area, which consisted of more than 60% tumor cells. Tissue sections were deparaffinized by the boiling method14 with incubation at 70℃ for 10 minutes and centrifugation for 10 minutes at maximum speed. DNA was extracted using a chelating ion-exchange resin (InstaGene Matrix, Bio-Rad, Hercules, CA, USA), and RNA was extracted using the High Pure RNA Paraffin Kit (Roche, Penzberg, Germany). After nucleic acid extraction, the purity and quantity of the nucleic acids were measured using a NanoDrop UV spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
Polymerase chain reaction
To investigate DNA integrity, PCR was performed on glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 60 bp), β-actin (148 bp), and human growth hormone (hGH; 434 bp). The primer sequences were as follows: GAPDH forward, 5'-ACACCCACTCCTCCACCTTT-3'; GAPDH reverse, 3'-TGACAAAGTGGTCGTTGAGG-3'; ACTB forward, 5'-CCGCCAGCTCACCATGGAT-3'; ACTB reverse, 5'-CACCATCACGCCCTGGTGC-3'; hGH forward, 5'-TGCCTTCCCAACCATTCCCTTA-3'; and hGH reverse, 5'-CCACTCACGGATTTCTGTTGTGTTTC-3'. All reactions were performed using the same cycling conditions: 95℃ for 30 seconds, 60℃ for 30 seconds, and 72℃ for 1 minute, for 35 amplification cycles. The PCR products were analyzed by electrophoresis on a 2% agarose gel.
Real-time reverse transcription-PCR
Real-time reverse transcription (RT)-PCR was performed to assess the integrity of the RNA. First, cDNA was synthesized using 1 µg total RNA, the Transcriptor First Strand cDNA Synthesis Kit, and oligo(dT) primer (Roche). Real-time TaqMan PCR for β-actin (97 bp) was then performed on each sample using the Universal ProbeLibrary (UPL) probe (Roche) and the Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The UPL probe sequences were as follows: left, CCAACCGCGAGAAGATGA; right, CCAGAGGCGTACAGGGATAG. Cycle parameters were 95℃ for 10 minutes and 50 cycles of 10 seconds at 95℃ and 60 seconds at 58℃.
Statistical analyses
The Mann-Whitney test or Kruskal-Wallis test was used to compare non-parametric continuous variables. Either the χ2 test or Fisher's exact test (two-sided) was performed to compare categorical variables. The trends of DNA PCR results according to the duration of decalcification were tested by linear-by-linear association. Results were considered significant when p-values were less than .05. All statistical analyses were conducted using the PASW ver. 19.0 (IBM Co., Armonk, NY, USA).
RESULTS
Comparison between formalin and methacarn
We compared the effects of formalin and methacarn fixation on DNA preservation by investigating 15 FFPE human cancer samples and 15 corresponding MFPE samples. For FFPE samples, surgical specimens were fixed with formalin for 24 to 48 hours. When GAPDH (60 bp), β-actin (148 bp), and hGH (434 bp) DNA fragments were amplified via PCR, all FFPE and MFPE DNA samples were positive for GAPDH and β-actin. Amplification of hGH was successful in all 15 MFPE samples but only in seven FFPE samples (p=.002). In most cases, a greater amount of PCR products was obtained from MFPE DNA than from FFPE DNA (Fig. 1).
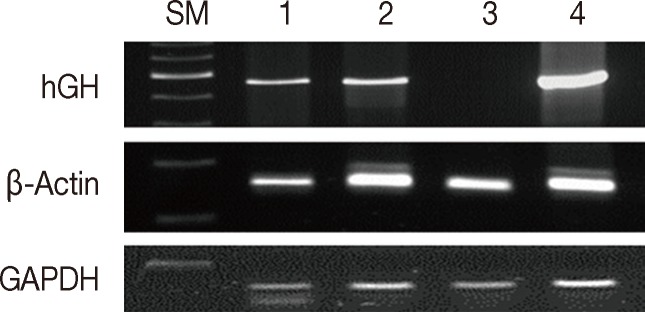
Comparison between formalin and methacarn fixatives by polymerase chain reaction of DNA of various sizes. Lanes 1 and 3, formalin fixation; lanes 2 and 4, methacarn fixation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 60 bp), β-actin (148 bp), and human growth hormone (hGH; 434 bp). SM, size marker.
Both RNA extraction and real-time RT-PCR of β-actin were performed using 12 FFPE and matched 12 MFPE human cancer samples, which were stored for less than 1 year. The yield was greater for FFPE samples than MFPE samples (637.87±312.17 ng/µL vs 149.43±84.75 ng/µL, p<.001). The purity, as measured by 260/280-nm ratios, was also higher for the FFPE samples than for the MFPE samples (1.97±0.06 ng/µL vs 1.85±0.18 ng/µL, p<.001). However, real-time RT-PCR was possible in all 12 MFPE samples but in nine of the 12 FFPE samples, which was statistically insignificant (p=.217). The Ct values were not significantly different between the FFPE and MFPE samples (p=.129, data not shown).
Effects of the duration of formalin fixation
Formalin fixation is the standard method used for routine tissue preservation. However, a standard duration of fixation has not been established, and duration of fixation may be prolonged to more than 48 hours because of holidays, more gross examination, or other circumstances. In order to evaluate the effects of the duration of formalin fixation, three human cancer samples were fixed in formalin for 3, 7, 30, 90, and 180 days at room temperature. As a control, tissue from the same samples was fixed in formalin for one day and then stored in 70% ethanol at 4℃ for 2, 6, 29, 89, and 179 days. The DNA integrity in the time-course treatment was assessed (Fig. 2), and there were considerable differences in the PCR results. Except for a weak band in the 3-day sample, amplification of hGH was unsuccessful in the samples with prolonged exposure to formalin. However, all samples kept in ethanol after 1-day formalin fixation had amplified hGH bands. Although the differences were not significant, the amount of GAPDH and β-actin PCR products decreased with increasing exposure to formalin. The DNA integrity was well preserved in the tissues kept in ethanol after 1-day formalin fixation.
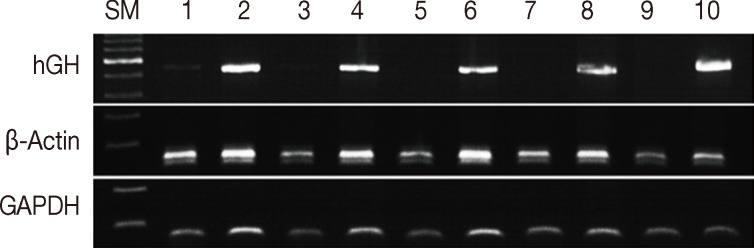
Assessment of DNA integrity according to the duration of formalin fixation. Lanes 1/3/5/7/9, fixed in formalin for 3/7/30/90/180 days at room temperature; lanes 2/4/6/8/10, fixed in formalin for one day and stored in ethanol at 4℃ for 2/6/29/79/179 days. SM, size marker; hGH, human growth hormone; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
The yield of RNA was greater for the samples fixed in formalin for 90 and 180 days than for the other samples (540.03±232.90 ng/µL vs 286.39±78.69 ng/µL, p=.026). However, prolonged exposure to formalin had no significant effect on the purity of RNA and the real-time RT-PCR products of β-actin (data not shown).
Effects of decalcification
To investigate the effects of decalcification, we decalcified five human cancer tissue samples for 0, 10, 30, 60, 120, and 180 minutes. The PCR results were significantly affected by the decalcification time. GAPDH was amplified in all five samples treated with decalcification solution for 0 minute, 10 minutes, and 60 minutes, but it was amplified in four samples decalcified for 30 minutes and 120 minutes and two samples for 180 minutes (p=.017) (Fig. 3A). β-Actin was amplified in all five control samples, four samples decalcified for 10 minutes, two samples for 30 minutes and 60 minutes, and none for 120 and 180 minutes (p<.001) (Fig. 3A). In contrast, hGH was amplified in four control samples, one sample for 10 minutes and 30 minutes, and none for 60, 120, and 180 minutes (p<.001) (Fig. 3A)
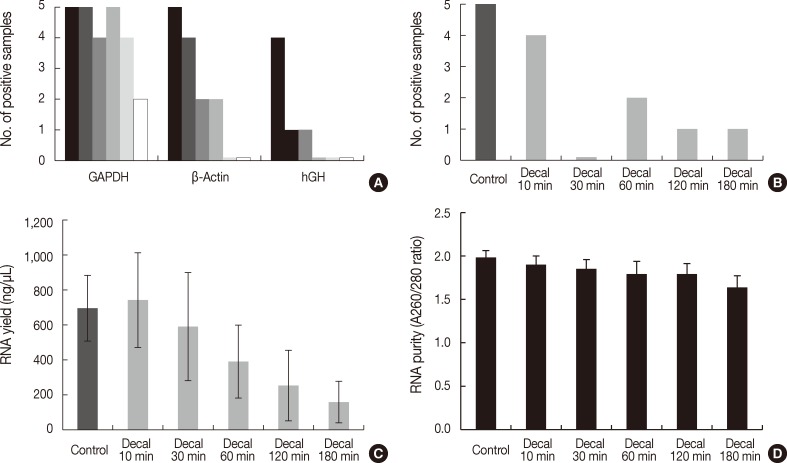
Bar graph representing polymerase chain reaction (PCR) results, reverse transcription (RT)-PCR results, the quantity of RNA, and the quality of RNA extracted from formalin-fixed paraffin-embedded tissues treated with decalcification solution. (A) PCR results of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), β-actin, and human growth hormone (hGH) according to duration of decalcification (0/10/30/60/120/180 minutes). (B) RT-PCR results of β-actin (97 bp). (C) RNA yield (ng/µL). (D) RNA purity (A260/280 ratios). Decal, decalcification.
Real-time RT-PCR of β-Actin was successful in four of five samples decalcified for 10 minutes, none for 30 minutes, two for 60 minutes, and one for 120 minutes and 180 minutes (p=.004) (Fig. 3B). The yield of RNA from decalcified tissues decreased gradually with increasing decalcification time (p=.004) (Fig. 3C). In addition, the purity (260/280 nm ratios) also decreased with increasing decalcification time (p=.009) (Fig. 3D).
Effects of the storage period
FFPE tissue stored at room temperature for 3 to 10 years has been used as an archival resource in many molecular studies. Thirty samples of human cancer tissue were randomly selected to investigate the effects of storage time on DNA preservation. All selected samples were from cases with enough cancer tissue for further experiments. These consisted of five samples stored for less than one month, five stored for six months, five stored for one year, five stored for three years, five stored for five years, and five stored for eight years. The PCR results were as follows; 13 out of 15 samples stored for 3 to 8 years were positive for GAPDH, 14 were positive for β-actin, and four were positive for hGH. Out of the 15 samples stored for ≤1 year, all 15 samples were positive for GAPDH and β-actin, and seven were positive for hGH. The PCR results were not significantly different between 3-8 year and ≤1 year storage groups (p>.05). The quantity and quality of the DNA did not significantly differ with the duration of storage (p>.05, data not shown).
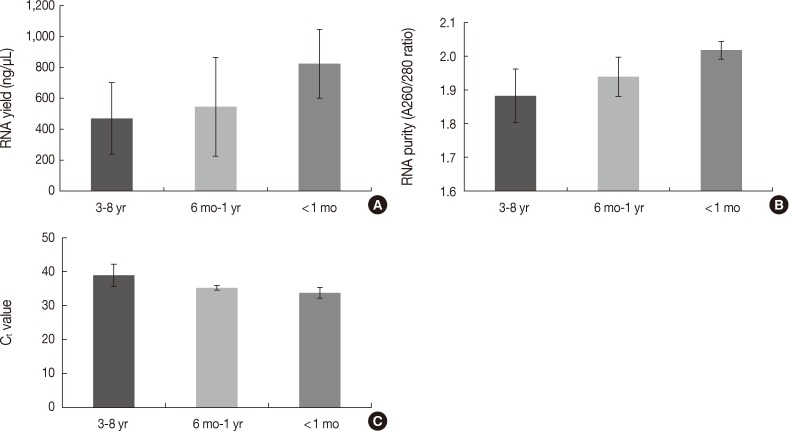
The quantity and quality of the RNA differed with the duration of storage. Tissues stored for longer periods had a smaller amount of RNA detected using the NanoDrop, although the difference was not statistically significant between 3-8 years and ≤1 year (469.86±231.87 ng/µL vs 637.87±312.17 ng/µL, p=.101) (Fig. 4A). Furthermore, there was a significant difference in RNA yield between >1 month and <1 month (499.90±264.97 ng/µL vs 823.66±223.06 ng/µL, p=.037). The purity also decreased with increasing storage periods (1.88±0.08 ng/µL for 3-8 years vs 1.97±0.06 ng/µL for ≤1 year, p=.008) (Fig. 4B). According to the real-time RT-PCR results, β-actin was amplified in seven of 12 samples (58.3%) with a 3-8 year storage time, five of eight samples (62.5%) with a one month to one year storage time, and all four samples (100%) with less than a one month storage time. Increasing the storage period increased the Ct value, which indicates fewer initial templates (37.83±1.18 for 3-8 years, 35.19±0.70 for one month to one year vs 33.73±1.60 for <1 month, p=.003) (Fig. 4C).
DISCUSSION
High-quality nucleic acid is important in molecular pathologic diagnosis, especially for cancer specimens. Accurate prediction of the prognosis and therapeutic response of cancer patients mainly depends on the sensitivity and specificity of the molecular diagnosis. However, nucleic acids from fixed tissues are reported to be easily damaged during pretreatment, which may lead to test failure or considerably different diagnostic results. Despite the previous studies showing nucleic acid damage during fixation and storage, the exact quality of extracted nucleic acids is still uncertain in daily practice. The fixation method and tissue storage should therefore be standardized to improve the quality of molecular pathologic diagnosis. In order to investigate the effects of fixatives, fixation method, and storage period, we tested the yield, purity, and integrity of nucleic acids from samples of gastrointestinal cancer tissue.
The fixatives affected the quality of extracted nucleic acids from human tissue samples. The formalin fixation method is currently the most common. However, the toxicity of formalin causes many problems.7 We compared formalin to methacarn with respect to nucleic acid integrity. The yield and purity of DNA from the FFPE samples did not differ from those of the MFPE samples (data not shown), and the yield and purity of RNA from the FFPE samples were higher than from the MFPE samples. hGH PCR was possible in all MFPE samples, but it was possible in only seven of the 15 corresponding FFPE samples. Furthermore, real-time RT-PCR for β-actin (97 bp) was possible in all MFPE samples but only in nine of the 12 FFPE samples. Therefore, the integrity of nucleic acids is likely to be greater in the MFPE samples than in the FFPE samples. Many pathologic laboratories employ a formalin-based fixation method. Formalin is a relatively good fixative for molecular diagnosis and FFPE samples are sufficiently high quality for PCR or RT-PCR of small-sized nucleic acids. However, considering the higher integrity of nucleic acids in the MFPE samples than in the FFPE samples, methacarn-based fixation may therefore be recommended to improve the integrity of nucleic acids.
Prolonged formalin fixation adversely affects the quality of tissue DNA, although it has only a minor effect on histopathology.11 The average size of DNA extracted from tissues fixed in buffered formalin decreases with increasing duration of fixation. Tissues fixed in buffered formalin for 3 to 6 hours yield greater amounts of high molecular-weight DNA.15 Srinivasan et al.7 recommended fixing tissue with buffered formalin for 3 to 6 hours to preserve nucleic acids. However, in practice tissues are routinely fixed for 24 to 48 hours, and sometimes the duration of fixation may be longer than 48 hours because of holidays or more sectioning. In this study, PCR of β-actin and GAPDH was successful in samples subjected to prolonged formalin fixation. However, PCR for larger DNA fragments (i.e., hGH, 434 bp) was not possible following prolonged formalin fixation. In addition, there were more β-actin and GAPDH PCR products in the samples subjected to 1-day formalin fixation that were stored in ethanol at 4℃ than in the samples subjected to prolonged formalin fixation. While PCR of small-sized DNA fragments is possible following prolonged formalin fixation, formalin fixation for one day and storage in ethanol is recommended to improve the quality of molecular diagnosis.
In various cancer specimens including papillary thyroid carcinoma, mucinous adenocarcinoma, as well as some bone and soft tissue tumor dystrophic calcification, is observed. Bone is also a common site of metastasis in various cancers (e.g., breast, prostate, thyroid, lung, and kidneys).16 The calcified tissue or bone specimens should be decalcified in order to obtain 4 to 8 µm thick FFPE tissue sections. Molecular pathology is also necessary in calcified tumor tissues for diagnosis and for determining a prognosis or predicting therapeutic responses. However, the degree of nucleic acid damage caused by decalcification is considered to be significant.12,17 To demonstrate the effects of a decalcification solution on tissue nucleic acids, we compared the yield, purity, and integrity of nucleic acids following various decalcification times. With a prolonged duration of decalcification, even small-sized DNA such as β-actin could not be amplified. Molecular diagnosis of calcified tissue should therefore be performed with caution, and nondecalcified paraffin blocks are recommended for accurate molecular diagnosis.
Most paraffin blocks are stored at room temperature. The rate of late recurrence in cancer patients has increased, and paraffin blocks that have been stored for more than five years are being used for molecular diagnosis.18 These archival paraffin blocks have also been a major source of many retrospective molecular studies. Archival specimens have shown great potential for use in PCR techniques.13,19 The present study demonstrated that small-sized DNA or RNA fragments were successfully amplified in most FFPE samples even if they were kept for several years at room temperature. However, recent specimens gave better PCR results than specimens that had been stored for several years. In particular, the yield, purity, and integrity of RNA by RT-PCR were better in the recent specimens. Real-time RT-PCR showed that β-actin was amplified in all samples (100%) with less than one month of storage time, but in 62.5% of samples stored for one month to one year, and 58.3% of samples stored for 3 to 8 years. Increasing the storage period also increased the Ct value, which was 37.83±1.18 for samples stored for 3 to 8 years, 35.19±0.70 for one month to one year, and 33.73±1.60 for <1 month. Therefore, the integrity of RNA is more affected by storage within the first year than for periods longer than one year. Some older samples could be used for real-time RT-PCR with optimal PCR product size, but the results of quantitative RT-PCR should be cautiously interpreted in the context of longer durations of storage. Although these results demonstrated a significant difference in the yield, purity, and integrity of RNA according to the storage period, this study is limited by a small sample size, and further large scale studies are needed.
In summary, we investigated the effects of fixatives, duration of formalin fixation, decalcification, and storage periods of paraffin blocks on the yield, purity, and integrity of nucleic acids from human cancer samples. Methacarn was better than formalin for nucleic acid preservation, especially high molecular-weight DNA. Prolonged formalin fixation negatively affected the integrity of DNA, but DNA quality could be preserved by storing the samples in ethanol at 4℃ following1-day formalin fixation. With decalcification, most nucleic acids were degraded, and PCR was unsuccessful. The yield, purity, and integrity of mRNA progressively decreased with prolonged storage of the paraffin blocks. Therefore, optimal fixatives and standardized procedures are essential for improving the quality of molecular pathologic diagnosis.
Notes
No potential conflict of interest relevant to this article was reported.