The proteomic landscape shows oncologic relevance in cystitis glandularis
Article information
Abstract
Background
The relationship between cystitis glandularis (CG) and bladder malignancy remains unclear.
Methods
We identified the oncologic significance of CG at the molecular level using liquid chromatography-tandem mass spectrometry-based proteomic analysis of 10 CG, 12 urothelial carcinoma (UC), and nine normal urothelium (NU) specimens. Differentially expressed proteins (DEPs) were identified based on an analysis of variance false discovery rate < 0.05, and their functional enrichment was analyzed using a network model, Gene Set Enrichment Analysis, and Gene Ontology annotation.
Results
We identified 9,890 proteins across all samples and 1,139 DEPs among the three entities. A substantial number of DEPs overlapped in CG/NU, distinct from UC. Interestingly, we found that a subset of DEP clusters (n = 53, 5%) was differentially expressed in NU but similarly between CG and UC. This “UC-like signature” was enriched for reactive oxygen species (ROS) and energy metabolism, growth and DNA repair, transport, motility, epithelial-mesenchymal transition, and cell survival. Using the top 10 shortlisted DEPs, including SOD2, PRKCD, CYCS, and HCLS1, we identified functional elements related to ROS metabolism, development, and transport using network analysis. The abundance of these four molecules in UC/CG than in NU was consistent with the oncologic functions in CG.
Conclusions
Using a proteomic approach, we identified a predominantly non-neoplastic landscape of CG, which was closer to NU than to UC. We also confirmed a small subset of common DEPs in UC and CG, suggesting that altered ROS metabolism might imply potential cancerous risks in CG.
Cystitis glandularis (CG), a common glandular lesion of the urinary bladder, is a cystic structure with a glandular lumen surrounded by outer urothelial cells that usually evolve from von Brunn’s nests after long-standing inflammation or irritation [1]. CG with intestinal metaplasia (IM) or the presence of mucin-containing goblet cells can produce abundant extracellular mucin that requires differentiation from primary adenocarcinoma, urachal adenocarcinoma, or metastatic adenocarcinoma from other organs [1,2]. CG can also develop a mass-like florid lesion that mimics bladder cancer on radiologic and cystoscopic examinations [3].
In addition to the clinical and histopathological disguise characteristics of CG, its relationship with malignancy has been discussed in studies with contrasting results [2]. For example, the co-occurrence of CG and adenocarcinoma has been frequently reported in the urinary bladder, which may indicate a connection between these two entities [1]. Furthermore, prior studies, using single gene or chromosome assays, underlaid the molecular basis supporting the premalignant nature of CG with or without IM by elucidating telomere shortening and chromosomal instability [4], p53 loss of heterozygosity and overexpression [5], and nuclear β-catenin expression [6]. In contrast, other studies failed to find a clear indication that CG, with or without IM, increased the future risk of developing bladder cancer during years of retrospective observation [7,8]. Comprehensive characterization would greatly help identify the pathobiology and clinical implications of CG.
In this study, we compared the proteomic landscape of CG with that of urothelial carcinoma (UC) and normal urothelium (NU). Differentially expressed proteins (DEPs) with functional enrichment were analyzed in CG, UC, and NU to identify the oncologic significance of CG at the molecular level.
MATERIALS AND METHODS
Patient selection
From the Seoul National University Hospital, 10 CG, 12 UC, and 9 NU specimens were collected after excluding patients who previously had any bladder tumor or intravesical treatment. Experienced pathologists (M.J. and H.S.R.) reviewed the diagnoses using hematoxylin-eosin slides based on the 2022 World Health Organization (WHO) classification [9]. All samples were obtained by transurethral resection of the urinary bladder, except for one NU, which was obtained from the ureter.
Liquid chromatography-tandem mass spectrometry
Formalin-fixed paraffin-embedded slides were scrapped and lysed in a sodium dodecyl sulfate-extraction buffer. After protein isolation, samples were sonicated and precipitated using acetone. Proteins were digested according to the filter-aided sample preparation procedure [10]. Tandem mass tag 6-plex labeling was employed for the peptide samples according to the manufacturer’s instructions, with modifications. After pooling the labeled peptides, the sample was separated into 12 fractions using Agilent 1290 bioinert high-pH reverse-phase liquid chromatography (Agilent, Santa Clara, CA, USA). For each peptide fraction, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was conducted using a Q-ExactivePlus mass spectrometer (Thermo Fisher, Waltham, MA) with an Ultimate 3000 RSLC system (Dionex, Sunnyvale, CA, USA), as previously described, with modifications [11]. Peptides were identified using a false discovery rate (FDR) of 1% as cutoff. Raw data are available in the ProteomeXchange Consortium (PXD027602) [12,13].
Proteomic data analysis
The MS raw data were processed using MaxQuant ver. 1.5.3.1 (Max Planck Institute of Biochemistry, Munich, Germany) [14] with the Andromeda engine [15]. For label-free quantification, the iBAQ algorithm was used on MaxQuant platform [16]. Using normalized abundance, we identified DEPs among CG, UC, and NU based on an analysis of variance (ANOVA) [14] test as a cutoff for permutation-based FDR < 0.05. Subsequently, the DEPs were hierarchically clustered based on Euclidean distance. Following a review of their abundance, we enlisted “UC-like” DEP clusters as “UC-like signature”, for which CG and UC overlapped but NU did not. All analyses were conducted using Perseus ver. 1.6.14.0 (Max Planck Institute of Biochemistry).
Functional enrichment analysis
To determine the biological processes represented by the lists of DEPs, we investigated protein-protein interactions (PPIs) using String database [17], Gene Ontology-biologic process (GOBP) annotation using Toppgene Suite [18], and the Molecular Signatures Database (MSigDB) Hallmark gene sets using pre-ranked Gene Set Enrichment Analysis (GSEA) [19]. To examine the associations among GOBPs, we reconstructed a network model using REVIGO [20]. Network models of PPI and GOBP were visualized using Cytoscape ver. 3.7.2 [21]. The z-scores of the selected proteins were compared between the group with and without IM using a two-tailed t-test.
RESULTS
We investigated 31 urothelial specimens, including 10 CGs, 12 UCs, and nine NUs, using a proteomic approach. A schematic outline of this study is shown in Fig. 1. The median ages were 40, 70, and 63.5 years, respectively, and the male-to-female ratios were 1.0, 0.4, and 0.9, respectively. Half of the specimens that contained CG were accompanied by IM. All UCs were confined to the mucosa (stage Ta), and eight (67%) were of WHO high grade.
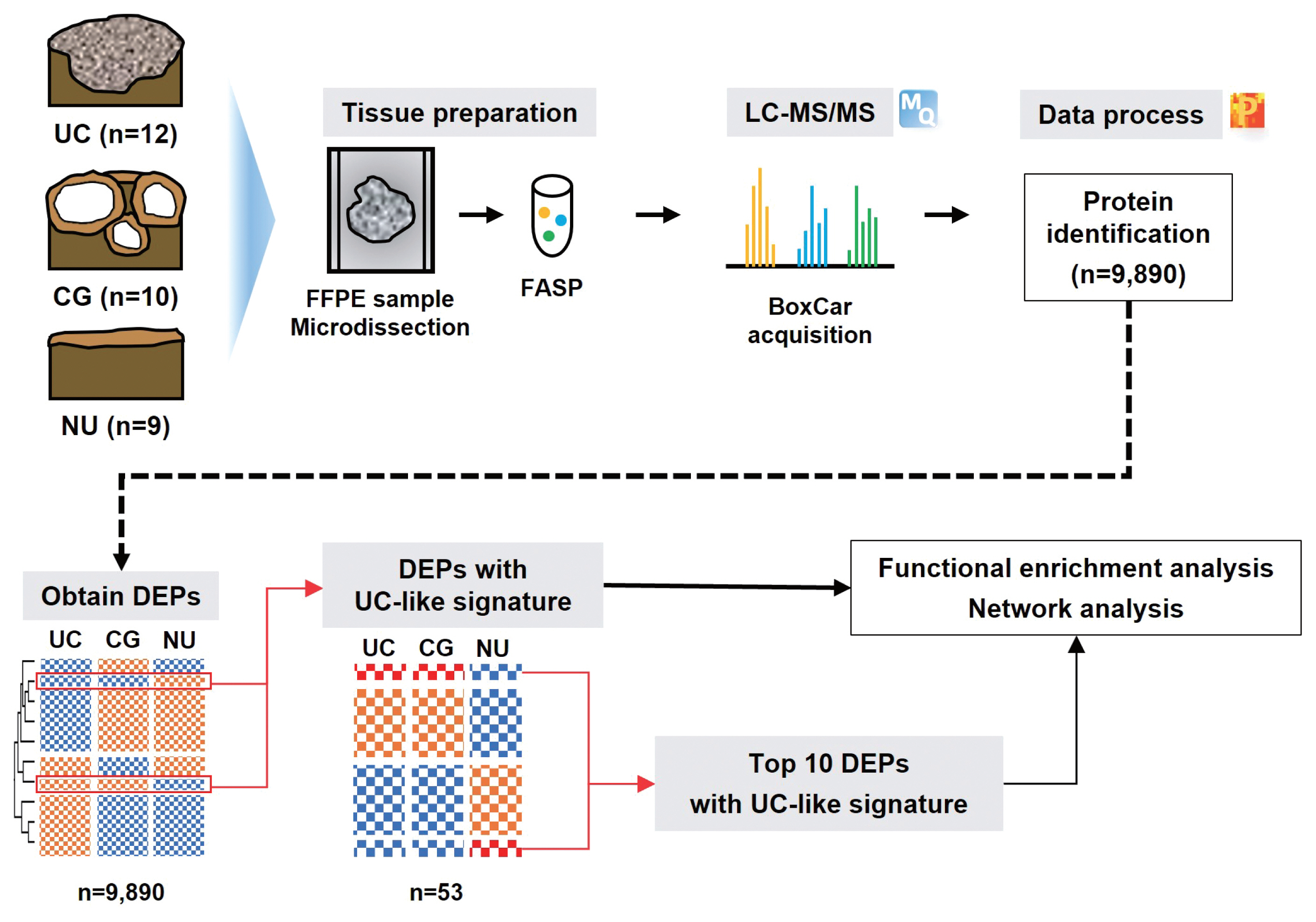
Schematic outline of the study. (Top) Liquid chromatography-tandem mass spectrometry (LC-MS/MS)–based proteomic analysis of 12 urothelial carcinoma (UC),10 cystitis glandularis (CGs), and nine normal urothelial (NU) specimens identified 9,890 proteins. Formalin-fixed paraffin-embedded (FFPE) slides were scrapped and proteins were isolated. After sonication and precipitation of samples, proteins were digested according to the filter-aided sample preparation (FASP) procedure. (Bottom) Differentially expressed proteins (DEPs) were discovered based on analysis of variance (false discovery rate < 0.05), and DEPs with UC-like signatures were revealed in CG. Functional enrichment analysis was performed for the DEPs and top 10 DEPs, respectively.
From LC-MS/MS analysis, we identified 9,890 proteins across all samples and 1,139 DEPs among the three entities (ANOVA FDR < 0.05). With unsupervised hierarchical clustering, a substantial number of DEPs were discovered to overlap with CG/NU in a way distinct from UC (Fig. 2). CG was not differentiated by the presence of IM (Fig. 2, asterisks). Interestingly, we found a subset of DEP clusters (n = 53, 5%) that were differentially expressed in NU but similarly expressed between CG and UC (Fig. 2, arrows); thus, these proteins were named as “UC-like signature” (Fig. 3A). Since the signature were similar within each group, the patterns of these signatures were thought to represent the general profile of CG.
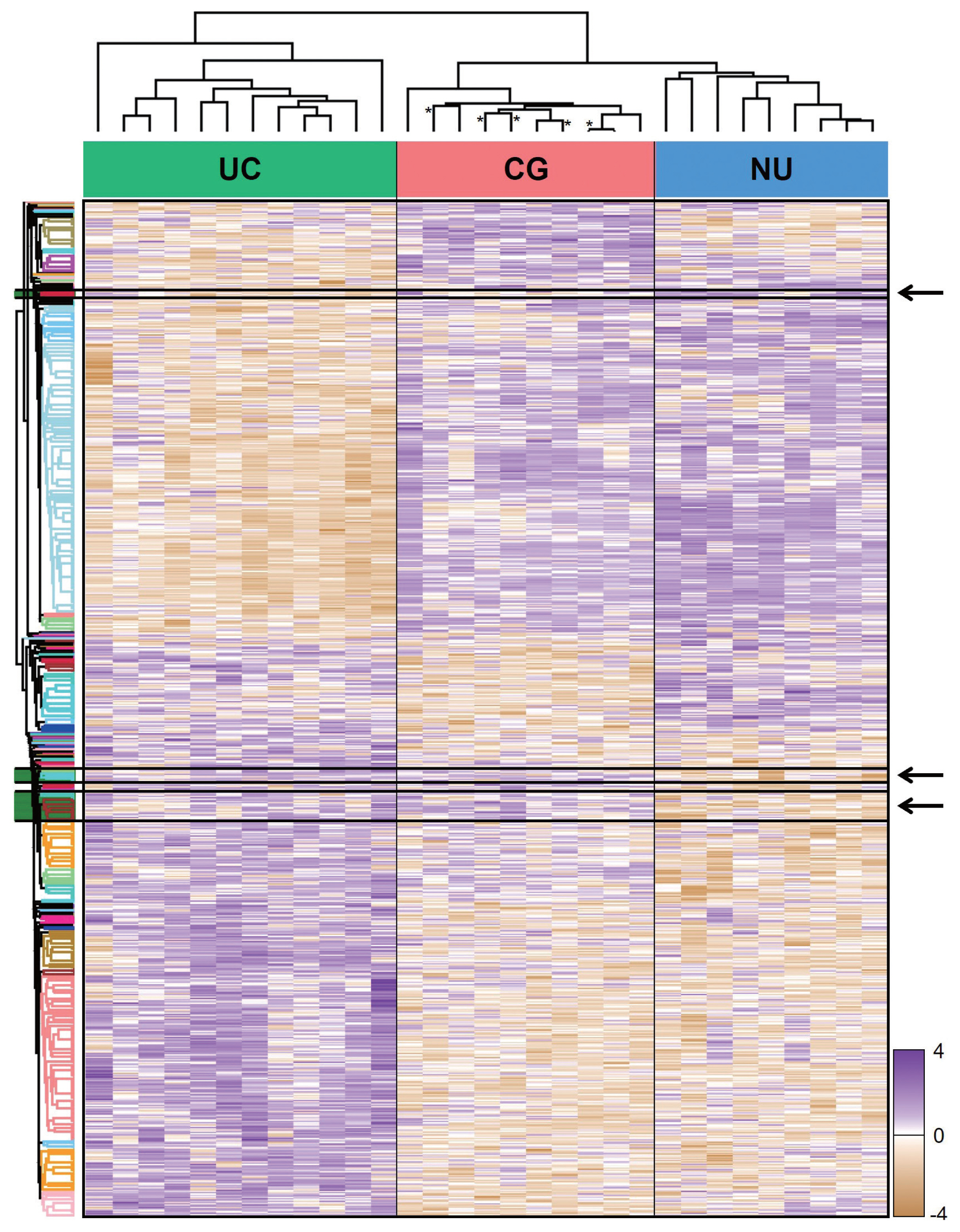
Unsupervised hierarchical clustering of 1,139 differentially expressed proteins identified by an analysis of variance test of urothelial carcinoma (UC), cystitis glandularis (CG), and normal urothelium (NU). Arrows indicate a “UC-like signature” that showed disparity between UC/CG and NU. Asterisks indicate CG samples showing intestinal metaplasia.
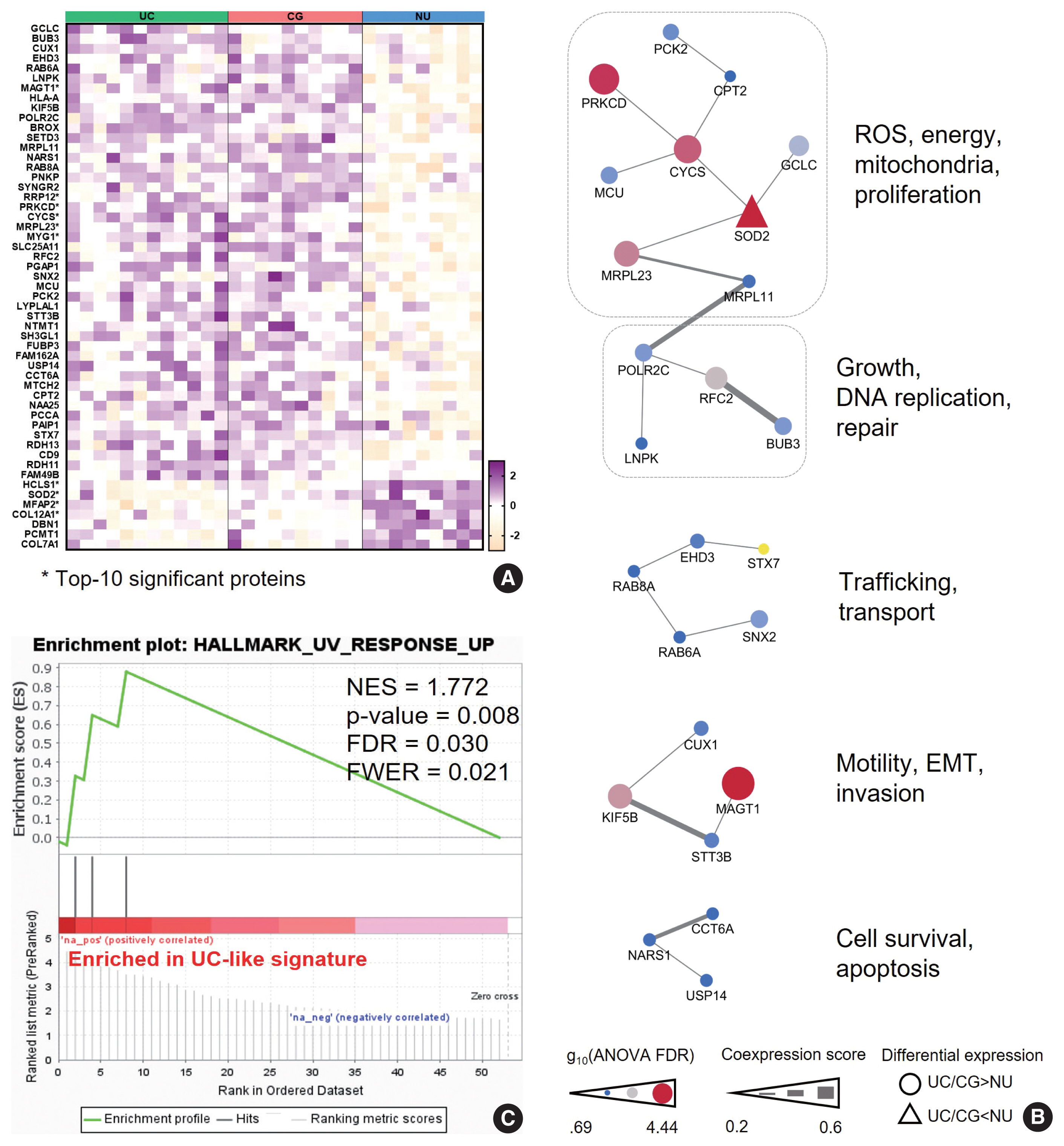
Pathobiologic characteristics of the “urothelial carcinoma (UC)-like signature”. (A) The UC-like signature proteins. Asterisks denote the 10 top-listed proteins by false discovery rate (FDR). (B) Protein-protein interaction networks of the “UC-like signature” proteins with their common functions. (C) Gene Set Enrichment Analysis of the “UC-like signature” shows enrichment of response to ultraviolet in molecular hallmark function. CG, cystitis glandularis; EMT, epithelial-mesenchymal transition; FWER, family wise error rate; NES, normalized enrichment score; NU, normal urothelium; ROS, reactive oxygen species.
We hypothesized that this signature might represent the UC-like characteristics of the CG. Using PPI analysis, we explored cellular processes represented by the “UC-like signature”. This resulted in sets of DEP networks within the “UC-like signature”, which were enriched for reactive oxygen species (ROS) and energy metabolism [22–24], growth and DNA repair [25,26], transport [27,28], motility and epithelial-mesenchymal transition (EMT) [29,30], and cell survival [31,32] (Fig. 3B). The unit associated with ROS and energy metabolism, which was connected to the one with growth and DNA repair functions, included the top-ranked DEPs (e.g., SOD2, PRKCD, CYCS, and MRPL23) (Fig. 3B). Using the official pre-ranked GSEA of the “UC-like signature”, we also identified significant enrichment of ultraviolet (UV) response (normalized enrichment score = 1.772, FDR = 0.03) for SOD2, PRKCD, and MRPL23 (Fig. 3C). UV radiation stimulates carcinogenic sequences by promoting ROS generation and DNA damage [33]. These data suggest that CG is predominantly a benign lesion, but oxidative stress might be a gateway theme representing oncogenic potential in CG.
To further characterize the ”UC-like signature”, we shortlisted the top 10 most significant DEPs (Fig. 3A, asterisk), followed by GOBP analysis (Fig. 4A). Subsequently, we constructed a network of connections between the GOBP terms. Consistent with the aforementioned single-protein networks, functional elements related to ROS metabolism, development, and transport were highlighted by GOBPs (Fig. 4B), which suggests that these functions could collectively substantiate the cancerous risks in CG.
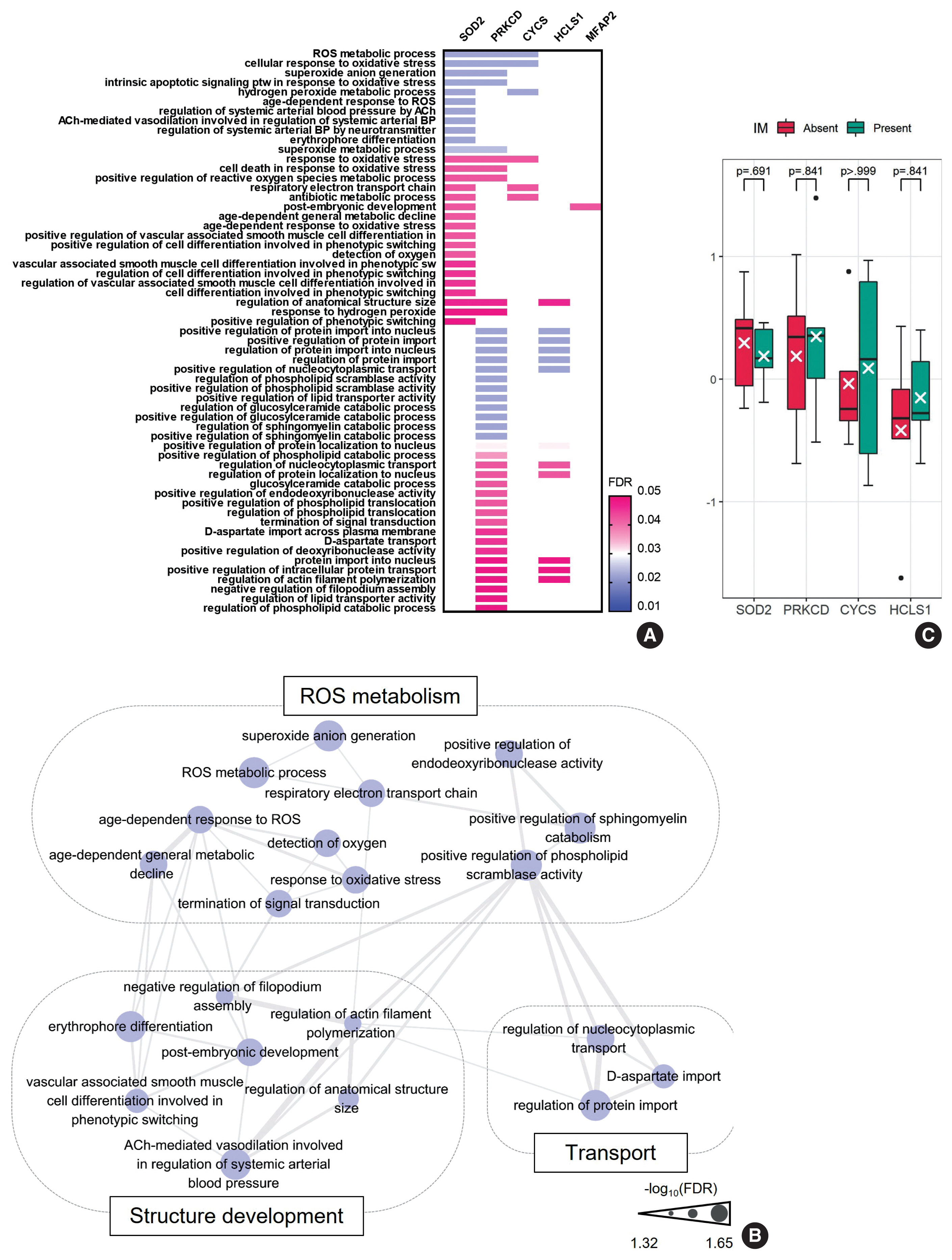
Imperative functions enriched in the “urothelial carcinoma-like signature”. (A) Gene Ontology-biologic processes (GOBPs) represented by the top 10 shortlisted proteins of “urothelial carcinoma-like signature”. (B) Network analysis of the GOBPs identifies reactive oxygen species (ROS) metabolism, structure development, and transport as common functional themes. (C) SOD2, PRKCD, CYCS, and HCLS1 were comparable between cystitis glandularis with and without intestinal metaplasia (IM). The value is expressed with z-scores of the proteome abundance. FDR, false discovery rate.
Of note, GOBPs were represented almost exclusively by four molecules, i.e., SOD2, PRKCD, CYCS, and HCLS1 (Fig. 4A), and their abundance in UC/CG compared to NU accordantly pointed toward oncologic functions. For example, previous studies have shown an association of decreased level of SOD2 and increased level of PRKCD, as observed in CG/UC versus NU, with ROS production [22,24]. Overt oxidative stress by activating oncogenic signaling pathways, DNA mutations, EMT, and stromal remodeling promotes bladder cancer development and progression [22]. In line with this, altered expression of CYCS and HCLS1 might deregulate oxidative respiration and the RAS signaling pathway, thereby enhancing the malignant behavior of bladder cancer [23,34]. We further examined whether these four statistically and functionally significant molecules were differentially expressed with IM and found no significant differences related to IM (Fig. 4C).
DISCUSSION
The malignant risk of CG and its association with bladder cancer have been disputed [1,2]. Considering the high prevalence of CG, there is a dire need to clarify its oncologic implications. Using LC-MS/MS, we revealed the proteomic landscape of CG in relation to malignant (UC) and normal (NU) urothelial tissues. CG generally presented an NU-like profile, in contrast to UC, indicating the overall benign nature of CG. By retrospectively observing a handful of patients with CG, consistent with this result, previous studies failed to confirm clear association between UC and CG [7,8]. Conversely, we confirmed that some of the clustered DEPs showed an opposite pattern across the diagnosis; these proteins were shared by CG and UC but not by NU. Using functional and network analyses, we found that these DEPs coded for ROS and energy metabolism, growth and DNA repair, transport, motility and EMT, and cell survival, further supporting the relation of this “UC-like signature” with the malignant risk in CG. To our knowledge, this is the first study to determine the oncogenic characteristics of CG using a comprehensive proteomic analysis. Further large-scale studies are required to determine the risks of UC development in patients with CG.
Using an interconnected functional network analysis of the shortlisted proteins, including SOD2, PRKCD, CYCS, and HCLS1, ROS metabolism, structural development, and transport functions were concordantly found to be significantly enriched in the “UC-like signature.” ROS are tightly regulated in normal cellular environments, and increased ROS act as versatile transducers for the generation and progression of bladder cancer [22]. For example, both UC and CG showed downregulation of SOD2, a scavenging molecule, and upregulation of PRKCD, an oxidative enzyme associated with ROS accumulation. ROS imbalance induces oxidative DNA damage, mutations, propagation of oncogenic signals including RAS, mitogen-activated protein kinases, phosphoinositide 3-kinase, and nuclear factor κB pathways, EMT, and stromal modification in bladder cancer [22,24]. Consistent with this, a previous study suggested that alterations in ROS metabolism participate in inflammation-associated cancer sequences [35,36]. In addition, upregulation of PRKCD can further support a malignant phenotype in bladder cancer by promoting migration and invasion [37]. CYCS is instrumental in regulating oxidative phosphorylation and energy metabolism, and high maintenance of CYCS, as identified in UC/CG, could induce metabolic modification in bladder cancer, or Warburg effect [23]. In addition, UC and CG showed lower HCLS1 levels than NU. HCLS1, an actin-binding molecule supporting cellular transport and trafficking, has been shown to result in adverse outcomes in bladder cancer [34,38]. We believe that the “UC-like signature” might reflect the oncogenic pathway related to altered ROS and energy metabolism and structural modification in CG, and the expression of these signature proteins deserves further study to determine the risk of aggression in CG.
This study lacked specimens with adenocarcinoma in the analysis. The malignant association of CG with IM has been conjectured, typically in terms of bladder adenocarcinoma, but definite evidence has not been confirmed [1]. Instead, altered ROS and energy metabolism have been implicated in the transformation of dysplasia to adenocarcinoma in the esophagus, consistent with the functions enriched in the “UC-like signature” [35]. In addition, the overall proteomic profile and biomarkers relevant to such functions showed no difference regardless of the presence of IM in CG. However, previous studies have suggested that CG with IM may be more advanced than CG without IM regarding tumorigenic potential, as exemplified by more robust telomere shortening or β-catenin activation [4,6]. Therefore, we reasonably speculate that the “UC-like signature” might reflect the malignancy risk encoded in CG irrespective of IM at a global level.
In conclusion, using comprehensive proteomic profiling, we identified a predominantly non-neoplastic landscape of CG that is closer to NU than to UC. Furthermore, we confirmed a small subset of common DEPs in UC and CG, suggesting altered functions of ROS metabolism that might imply potential cancerous risks in CG.
Notes
Ethics Statement
All procedures performed in the current study were approved by the Institutional Review Board (IRB) (H-2009-163-1160; 05 October 2020) per the 1964 Helsinki Declaration and its later amendments. Formal written informed consent was not required, with a waiver by the appropriate IRB.
Availability of Data and Material
The datasets generated or analyzed during the current study are available in the ProteomeXchange Consortium (http://www.proteomexchange.org/) via the PRIDE database (accession: PXD027602).
Code Availability
Not applicable.
Author Contributions
Conceptualization: HSR. Data curation: HSR. Formal analysis: JYK, MJ, HSR. Funding acquisition: MJ. Investigation: JYK, MJ, HSR. Methodology: HSR. Project administration: HSR. Resources: HSR. Software: DH, HK, HSR. Supervision: HSR. Validation: DH, HK, MJ, HSR. Visualization: JYK, MJ, HSR. Writing—original draft: JYK, MJ. Writing—review and editing: MJ, HSR.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
This research was supported by the Korean Society of Pathologists Grant (No. KSPG2020-04).
