Biomarker testing of cytology specimens in personalized medicine for lung cancer patients
Article information
Abstract
Every patient with advanced non–small cell lung cancer (NSCLC) should be tested for targetable driver mutations and gene arrangements that may open avenues for targeted therapy. As most patients with NSCLC in the advanced stage of the disease are not candidates for surgery, these tests have to be performed on small biopsies or cytology samples. A growing number of other genetic changes with targetable mutations may be treatable in the near future. To identify patients who might benefit from novel targeted therapy, relevant markers should be tested in an appropriate context. In addition, immunotherapy of lung cancer is guided by the status of programmed death-ligand 1 expression in tumor cells. The variety and versatility of cytological specimen preparations offer significant advantages for molecular testing; however, they frequently remain underused. Therefore, evaluating the utility and adequacy of cytologic specimens is important, not only from a lung cancer diagnosis, but also for the large number of ancillary studies that are necessary to provide appropriate clinical management. A large proportion of lung cancers is diagnosed by aspiration or exfoliative cytology specimens; thus, optimizing strategies to triage and best use the tissue for diagnosis and biomarker studies forms a critical component of lung cancer management. In this review, we discuss the opportunities and challenges of using cytologic specimens for biomarker testing of lung cancer and the role of cytopathology in the molecular era.
The genomic landscape of non–small cell lung carcinoma (NSCLC) is constantly evolving, with the discovery of a growing number of molecular alterations and associated targeted therapies that have a huge impact on patient care. The College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology (CAP/IASLC/AMP) issued a guideline in 2013 to provide a roadmap for molecular testing to select patients for treatment with targeted tyrosine kinase inhibitors [1]. However, since 2013, many new emerging target molecules have been identified, including mutations in BRAF, ERBB2, and in MET exon 14, and rearrangements in RET. The guidelines were thus updated in 2018 and endorsed by the American Society of Clinical Oncology [2]. The latest version of the molecular testing guidelines for NSCLC recommends that molecular studies be performed before any systemic therapy is administered to assess a minimum of epidermal growth factor receptor (EGFR) mutations, anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) rearrangements, and BRAF mutations [2]. In addition, with the advent of immunotherapy, evaluating the expression level of programmed death-ligand 1 (PD-L1) has been recommended for the identification of patients who respond to immune checkpoint inhibitors [3].
Despite the rapid increase in the number of clinically relevant biomarkers for advanced-stage NSCLC, limited availability of tissue samples for molecular analysis remains a major challenge. Undoubtedly, both surgical and biopsy samples still represent the “gold standard” of the starting material for molecular purposes. This is mainly because formalin-fixed and paraffin-embedded (FFPE) histological specimens have the advantage of enabling morphological evaluation, and do not require additional molecular validation. However, in real-world clinical practice, obtaining sufficient tissue specimens from advanced-stage NSCLC patients is highly impractical. In this setting, cytological samples may be an excellent alternative to histological samples. The updated CAP/IASLC/AMP guidelines recommend the adoption of cytological smears for molecular analysis of advanced-stage NSCLC patients [2].
Here, we critically reviewed the molecular cytopathology of NSCLC, including (1) the various types of lung cytology specimens, preparation methods, and preanalytic factors affecting nucleic acid yield and downstream biomarker testing; (2) the variety of molecular techniques applied to cytology samples; and (3) the opportunities and challenges in biomarker testing of cytological specimens.
WHICH CYTOLOGICAL SPECIMENS CAN BE USED?
The most common cytologic sampling methods in NSCLC cancer patients are fine needle aspiration of computed tomography–guided or electromagnetic navigation bronchoscopy–guided lung lesions and endobronchial ultrasound–guided lymph nodes and collection of exfoliative samples such as body fluid/effusions, bronchial brushing/washings, bronchoalveolar lavages, and sputum. Occasionally, minimally invasive aspiration samples from distant, deep-seated, or superficial metastatic lesions are also included. Cytological preparations that can be used for molecular studies include cell blocks (CBs), needle rinses, direct smears, cytospins, and liquid-based preparations (LBPs). To provide the best material for biomarker testing, the correct choice among different cytological preparations of the same sample should be considered. Representative microscopic images and advantages/ disadvantages of different cytological preparations are shown in Table 1 and Fig. 1, respectively.
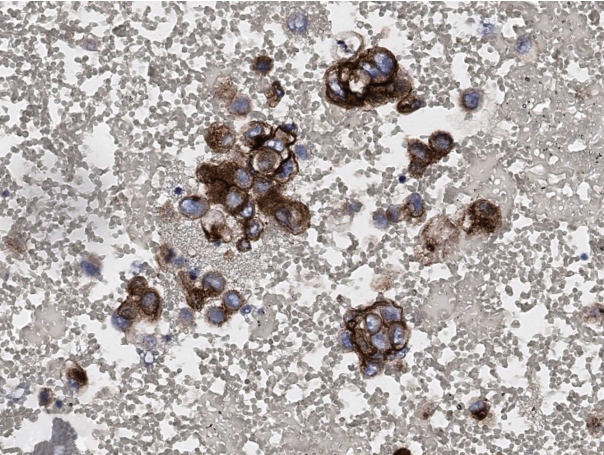
Reperesentatvie microscopic findings according to cytologic preparations diagnosed as metastatic non-small cell carcinoma. (A) Direct smear of endobronchial ultrasound–guided fine needle aspiration samples from mediastinal lymph nodes (Papanicolaou stain). (B, C) Cytospin and liquid based preparation of pleural fluids from advanced lung cancer patients, respectively (Papanicolaou stain). (D) Cell block from (B).
CBs are most commonly used for molecular diagnostic testing because they closely recapitulate FFPE specimens and generally do not require further validation; in addition, it is relatively easy to acquire multiple serial sections to perform immunocytochemical and molecular diagnostic assays [4]. However, on-site adequacy evaluation cannot be performed on CB, which leads to unpredictable results of cellularity and sometimes renders the CB paucicellular. Additionally, tumor cells are often widely spaced, resulting in low tumor cellularity per section area. In addition, the standard 4–5-μm CB sections do not represent the entire nuclei from the cell and are likely to have lower nucleic acid yields for molecular testing per cell than the whole cells obtained from other non–formalin-fixed cytologic preparations. To increase nucleic acid yield, not only providing more sections, but also macrodissecting the regions of highest tumor cellularity may be an option [5].
Direct smears and cytospins that are either air-dried or ethanol-fixed are not formalin-fixed preparations, which have the obvious advantage of obtaining an excellent quality material with a higher nucleic acid yield than CBs [6,7]. Besides being suitable for DNA-based next-generation sequencing (NGS) analysis, direct smears may also be appropriate for RNA-based NGS testing [8]. In addition, they offer the advantage of on-site adequacy assessment and better triaging of the sample for diagnosis and ancillary studies. In cases in which all or most of the diagnostic material is on a single smear/cytospin preparation that will be used for biomarker testing, CAP guidelines allow for the sacrifice of diagnostic material when medically necessary; the diagnostic slide can be digitally scanned for the archives to mitigate the medicolegal constraints [9].
Finally, LBPs represent a valuable alternative to conventional preparations to avoid inadequate management of the achieved material. The advantages of liquid-based cytology (LBC) specimens include optimal cell preservation, easy specimen transportation because of the stability of cells at room temperature, and minimal background debris and blood on slides [10-12]. Nucleic acid can be extracted from both rinse solutions, and cells can be scraped off the sides [13-15]. Of note, the properties of the different preservative solutions used in LBC may affect downstream molecular analysis. Some studies have indicated that cells preserved in CytoLyt (Cytyc Corp., Boxborough, MA, USA) solution provide higher DNA yields than those preserved in CytoRich Red fluid [16]. One study comparing cellularity and DNA yield between ThinPrep (Cytyc Corp.) slides (CytoLyt LBC) and direct smears reported greater cellularity and significantly higher average DNA yields in the latter [13], whereas a more recent study reported issues with long-term DNA stability and accelerated DNA degradation in LBC samples when compared with conventional smears [17].
WHAT TYPES OF BIOMARKER TESTING CAN BE PERFORMED ON CYTOLOGY SPECIMENS?
Polymerase chain reaction–based tests
Molecular testing for genetic mutations, such as in EGFR, in cytologic specimens has been described using a variety of polymerase chain reaction (PCR)–based techniques, including direct sequencing, real-time PCR, pyrosequencing, and peptide nucleic acid–locked nucleic acid [14,18-21]. Different techniques have different limits of detection and reference ranges, and the choice of platform used for the detection of mutations remains a decision of the individual molecular laboratories performing the assay (Table 2). Although the CAP/IASLC/AMP guidelines recommend a technique used to detect mutations in specimens with >50% tumor fraction [2], more sensitive platforms capable of detecting mutations in specimens with <10% tumor are strongly encouraged. The adequacy of cytological samples for mutational analysis is another important factor that is assessed according to tumor cellularity and viability. The CAP/IASLC/AMP guidelines recommend testing from samples with as little as 20% tumor cellularity because current mutation testing uses PCR-based methods that are more sensitive than unmodified Sanger sequencing [2]. In our study for the detection of EGFR mutation using the cytologic samples, the following parameters were correlated with the most reliable EGFR mutation results using the pyrosequencing method (100% concordance with the corresponding histologic specimens) in cytologic samples: a DNA concentration >25 ng/μL, content of >30 tumor cells, or a tumor percentage >30% [22].
Fluorescence in situ hybridization
To detect gene rearrangements such as in ALK and ROS1, fluorescence in situ hybridization (FISH), which was verified as a break-apart probe in a clinical trial, was first certified as a companion diagnostic test [23]. Previously, FISH testing was recommended only for CBs, but the 2018 CAP/IASLC/AMP guidelines recommend the use of conventional cytologic preparations for FISH [2]. Several groups have reported the potential and usefulness of the probe in non-formalin cytological preparations, including Diff-Quik and Papanicolaou-stained smears, as well as LBC ThinPrep slides; some report better performance than that seen with CB sections [24-26]. The advantage of using smears or LBCs is that whole-cell nuclei are analyzed to eliminate signal loss due to truncating artifacts, as seen in FFPE sections, but the disadvantage is that thresholds for positive and negative cutoffs are established using FFPE histological materials [27]. Therefore, independent standardization and validation of each sample type are required.
Immunocytochemistry
After the ALK D5F3 CDx Assay (Ventana Medical Systems, Tucson, AZ, USA) was approved by the Food and Drug Administration (FDA), immunohistochemistry (IHC) has been established as a confirmatory diagnostic test rather than screening, supplementing the shortcomings of FISH in detecting ALK rearrangement. The 2018 CAP/IASLC/AMP guidelines recommend ALK IHC as a valid alternative to the FISH (Fig. 2) [2]. The FDA has approved the assay only for “routinely processed, paraffin-embedded specimens fixed in neutral-buffered formalin.” However, several studies have demonstrated the feasibility of ALK immunocytochemistry (ICC) for direct smears and LBPs [28,29]. The updated guidelines recommend using ROS1 IHC with D4D6 (Cell Signaling Technology, Danvers, MA, USA) only as a screening test that requires confirmation by a molecular or cytogenetic method [2]. A limited number of studies using ROS1 FISH in cytological specimens are available [30,31]. Studies on the use of ROS1 ICC in cytology preparations are currently limited in the literature [32,33].
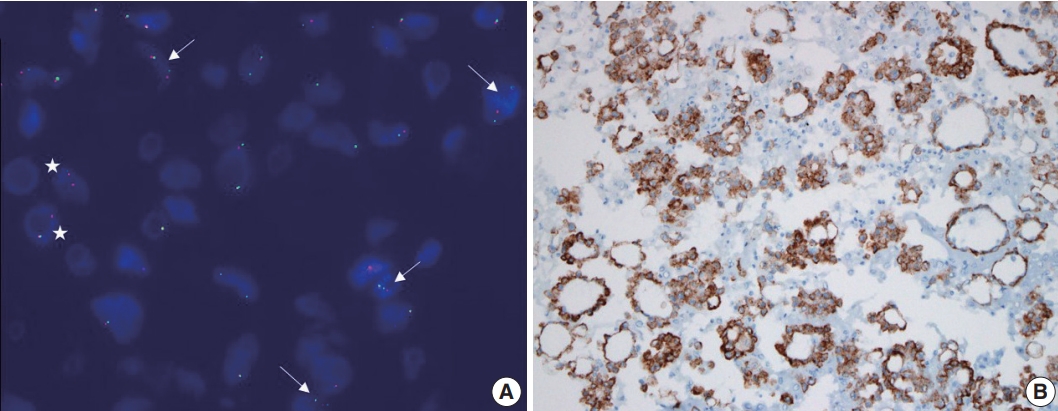
Fluorescence in situ hybridization using an LSI anaplastic lymphoma kinase (ALK) dual-color break-apart probe (A) and immunocytochemical staining using ALK D5F3 clone (B) on cytologic blocks of lung adenocarcinoma. (A) Two distinct red and green (break apart) signals with one intact fusion signal patterns (arrows) and an isolated red signal (IRS) with one intact fusion signal pattens (asterisks) were observed in > 50% of tumor cells. (B) Tumor cells exhibited strong, granular, and diffuse cytoplasmic signal, indicating aberrant ALK protein expression generated by gene fusion.
However, application of these assays to cytologic specimens requires meticulous validation because these assays are validated primarily on FFPE histological tissue samples. The lack of standardized processing protocols in cytology lead to a variety of preanalytic variables that can affect the antigenicity of antibodies used for predictive biomarker testing. CBs are most widely used for ICC; however, there is no standardized protocol for the type of collection media, pre-fixation, and processing techniques, and there is wide variation among pathology laboratories. Several recent studies have highlighted issues with immunostaining of specific markers that demonstrate reduced antigenicity and false-negative results, mostly related to ethanol or methanol-based fixatives used prior to CB preparation [34,35]. Non-CB cytological preparations present an even greater challenge for ICC validation. Immunostaining of ethanol-fixed smears or cytospins is used more frequently, with prior Papanicolau staining that can identify areas or cells of interest, or air-dried unfixed extra slides that can be used for ICC, usually after post-fixation step involving formalin or acetone [36,37]. In a recent meta-analysis of ALK ICC, the smear showed a slightly lower sensitivity than that of CB. These results are interpreted to indicate that the expression intensity of the antibody is low in alcohol-fixed smear slides because the expression of the antibody is optimized in FFPE [37].
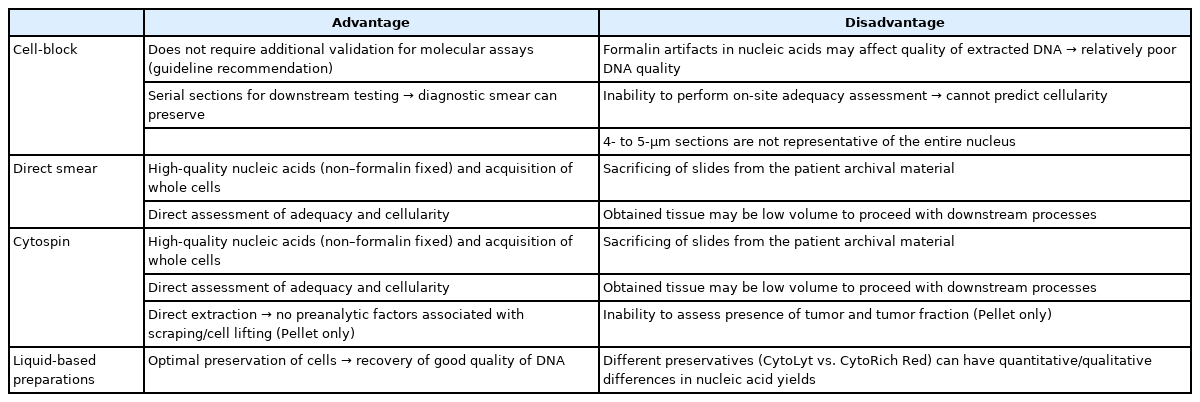
Unfortunately, guidelines for PD-L1 testing have not yet been provided even in updated guideline [2,38]. Although cytology specimens were not included in the initial clinical validation studies for PD-L1, several groups have evaluated the feasibility of PD-L1 in cytology specimens and have demonstrated results that are comparable to those of paired histologic samples [39,40]. Fig. 3 shows representative microscopic findings of strong positive expression of PD-L1 stained in CB of a patient diagnosed with metastatic adenocarcinoma in pericardial fluid. Lozano et al. reported the variation in patterns of PD-L1 expression on cytological specimens; because entire cells were present on direct smear, tumor cells often demonstrated a folded cell membrane, demonstrating a thick and strong membranous positivity [41]. Taken together, ICC in cytologic specimens remains a number of challenges to be solved throughout the standardization of protocols that can control preanalytical variables, rigorous validation of staining results, and systematic training for interpretation.
Next-generation sequencing
Next-generation sequencing (NGS) is a fascinating tool that can analyze multiple genetic alterations simultaneously, even when applied to cytological samples with low DNA/RNA yields. The advantages of using cytology specimens for NGS include quicker fixation or, if the platform is validated, minimal/no fixation, improving the quality of the input nucleic acids. Several studies using cytological material, including CBs as well as non-FFPE substrates, have shown them to be equally effective in the genomic profiling of NSCLC by NGS analysis [42-46]. In fact, some studies have indicated better quality metrics when comparing NGS analysis in non-FFPE cytologic substrates versus FFPE materials [6,47]. However, studies of the application of NGS to cytology specimens generally have a retrospective design, and only samples characterized by at least 20% of tumor cells, which may not fully reflect current practice, were selected. Therefore, it is crucial to establish the minimum number of cells needed to allow an NGS approach from cytology sample in routine practice. In any case, sample requirement depends on target capture, gene panel, and platform types. Illumina NGS usually requires more cells and/or higher DNA input than Ion Torrent NGS; thus, the latter seems to be more efficient with the cytopathologist specimens [5]. Recently, it was shown that lowering the input DNA concentration below the manufacturer’s recommended threshold of 10 ng (>0.8 ng/μL) is feasible leading to a marked increase in the NGS success rate from 58.6% to 89.8% [5,48]. More important than DNA input is the percentage of neoplastic cells. The preferential amplification of a small number of DNA in a small amount of cancer cells may only be representative of non-neoplastic components, which may lead to false-negative results. Macrodissection or microdissection are especially important for enrichment of viable tumor cells [5,49].
CHALLENGES AND FUTURE DIRECTIONS
In this era of personalized medicine, biomarker testing of cytology preparations is a relatively new and rapidly developing field with great potential, especially in patients with advanced NSCLC. However, cytological specimens continue to be excluded from most biomarker-driven clinical trials, primarily because of the failure to exploit the variety of different specimen preparations and the lack of validation for different assays. The lack of standardization of specimen processing among laboratories is major limitation. Therefore, the implementation of strategies to optimize and standardize procedures for specimen acquisition, processing, and tissue extraction is critical to maximize the use of cytological samples for ancillary studies and to provide relevant information for inclusion in clinical trial design. At minimum, confirmation of validation for cytology preparations and close check of quantity and quality of submitted material is also expected.
CONCLUSION
In conclusion, biomarker testing can be used for a variety of cytologic specimen types and preparations. This is of utmost importance for NSCLC patients, where the cytology specimen may be the only sample available for diagnosis and ancillary studies. Therefore, a thorough understanding of the potential and the limitations of these substrates is required to properly classify and use them for molecular studies that can guide patient management.
Notes
Ethics Statement
Not applicable.
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Author contributions
Conceptualization: HK, JHC. Writing—original draft: HK. Writing—review & editing: HK, JHC. Approval of final manuscript: all authors.
Conflicts of Interest
JHC, a contributing editor of the Journal of Pathology and Translational Medicine, was not involved in the editorial evaluation or decision to publish.
Funding Statement
No funding to declare.