Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 50(5); 2016 > Article
-
Original Article
SIRT7, H3K18ac, and ELK4 Immunohistochemical Expression in Hepatocellular Carcinoma - Hye Seung Lee, Wonkyung Jung, Eunjung Lee1, Hyeyoon Chang, Jin Hyuk Choi, Han Gyeom Kim, Aeree Kim, Baek-hui Kim,
-
Journal of Pathology and Translational Medicine 2016;50(5):337-344.
DOI: https://doi.org/10.4132/jptm.2016.05.20
Published online: August 5, 2016
Department of Pathology, Korea University Guro Hospital, Seoul, Korea
1Department of Pathology, Korea University Anam Hospital, Seoul, Korea
- Corresponding Author Baek-hui Kim, MD, PhD Department of Pathology, Korea University Guro Hospital, Korea University School of Medicine, 148 Gurodong-ro, Guro-gu, Seoul 08308, Korea Tel: +82-2-2626-1472 Fax: +82-2-2626-1486 E-mail: 'maelstrom@naver.com'
• Received: March 16, 2016 • Revised: May 18, 2016 • Accepted: May 19, 2016
© 2016 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- SIRT7 is one of the histone deacetylases and is NAD-dependent. It forms a complex with ETS-like transcription factor 4 (ELK4), which deacetylates H3K18ac and works as a transcriptional suppressor. Overexpression of SIRT7 and deacetylation of H3K18ac have been shown to be associated with aggressive clinical behavior in some cancers, including hepatocellular carcinoma (HCC). The present study investigated the immunohistochemical expression of SIRT7, H3K18ac, and ELK4 in hepatocellular carcinoma.
-
Methods
- A total of 278 HCC patients were enrolled in this study. Tissue microarray blocks were made from existing paraffin-embedded blocks. Immunohistochemical expressions of SIRT7, H3K18ac and ELK4 were scored and analyzed.
-
Results
- High SIRT7 (p = .034), high H3K18ac (p = .001), and low ELK4 (p = .021) groups were associated with poor outcomes. Age < 65 years (p = .028), tumor size ≥ 5 cm (p = .001), presence of vascular emboli (p = .003), involvement of surgical margin (p = .001), and high American Joint Committee on Cancer stage (III&V) (p < .001) were correlated with worse prognoses. In multivariate analysis, H3K18ac (p = .001) and ELK4 (p = .015) were the significant independent prognostic factors.
-
Conclusions
- High SIRT7 expression with poor overall survival implies that deacetylation of H3K18ac contributes to progression of HCC. High H3K18ac expression with poor prognosis is predicted due to a compensation mechanism. In addition, high ELK4 expression with good prognosis suggests another role of ELK4 as a tumor suppressor beyond SIRT7’s helper. In conclusion, we could assume that the H3K18ac deacetylation pathway is influenced by many other factors.
- Case selection and review
- A total of 278 HCC patients, who underwent curative surgery at Korea University Guro Hospital between 2000 and 2013 were enrolled in this study. All hematoxylin and eosin–stained slides were reviewed. To classify the pathologic staging, the American Joint Committee on Cancer (AJCC) staging system (seventh edition) was applied [16]. Tumor differentiation was graded according to the Edmondson-Steiner grading system [17]. Tumor necrosis was evaluated by gross photo and microscopic examination. The results were categorized as ‘present’ when necrosis was detected, even in minor amounts. Missing data were excluded, and clinicopathologic data including sex, age and TNM stage were obtained from patients’ medical records. This study was approved by the Institutional Review Board of Korea University Guro Hospital (KUGH14140-001).
- Tissue microarray construction and immunohistochemistry
- Representative tumor areas in all patients (n=278), and nontumor areas in 52 patients were obtained and embedded on paraffin blocks. Tissue cores 2.0 mm in diameter were collected from paraffin-embedded donor blocks and embedded in tissue microarray (TMA) blocks. IHC staining was performed on 4 μm TMA section slides using Bond-Max auto-stainer (Leica, Wetzlar, Germany). After incubation with primary antibodies, Bond Polymer Refine Detection kit (Leica) was used for chromogenic reaction. The following primary antibodies were used in this study; ELK4 (1:50, rabbit polyclonal, Novus, Littleton, CO, USA), SIRT7 (1:100, rabbit polyclonal, Abcam, Cambridge, MA, USA), and H3K18ac (1:1000, rabbit polyclonal, Abcam).
- IHC assessment
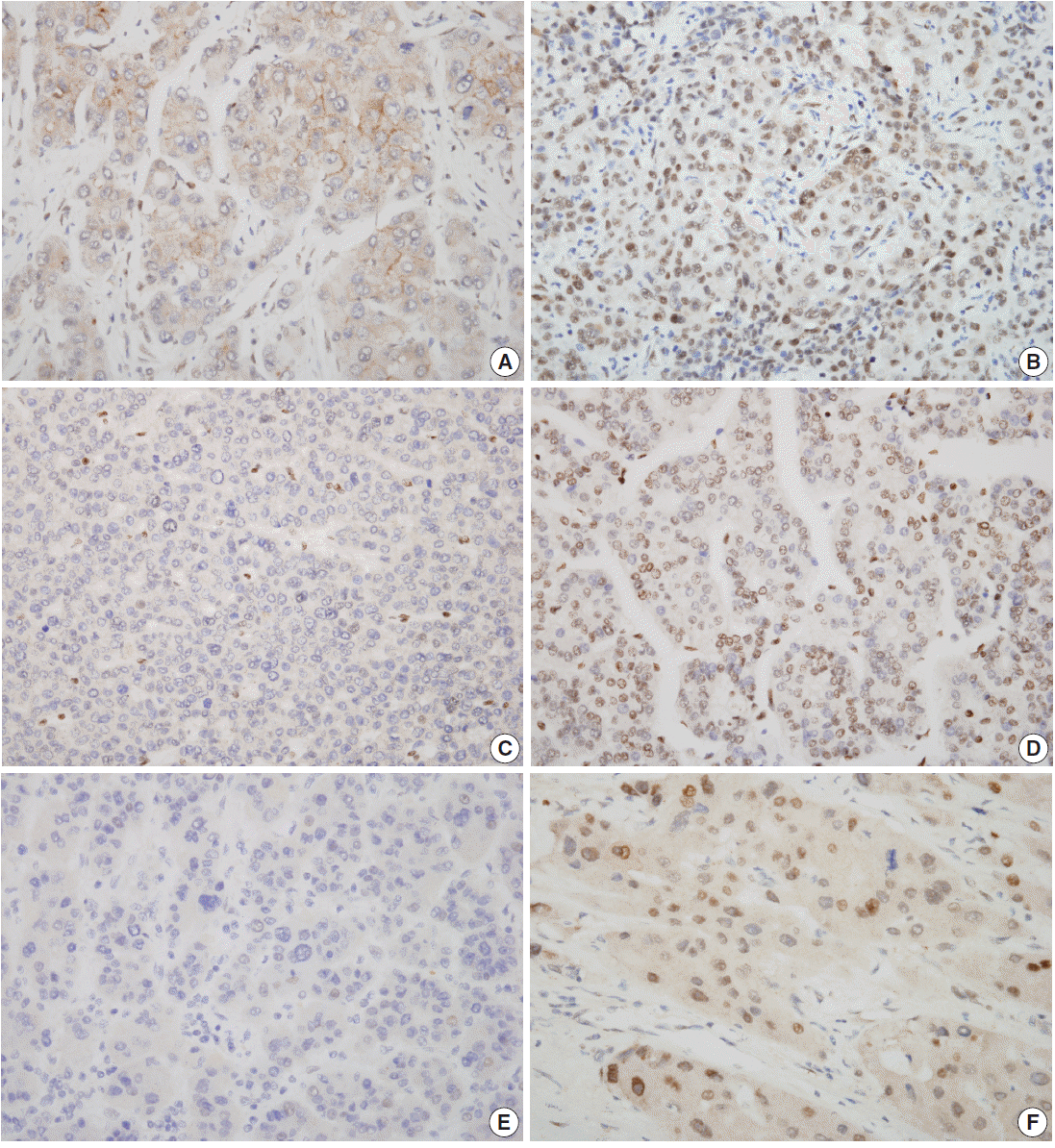
- Nuclear staining patterns of tumor cells for ELK4, SIRT7, and H3K18ac were analyzed (Fig. 1). More than 10% of stained tumor cells were categorized as positive. Scoring of intensity was done according to the following scale: 0, no or equivocal; 1, weak; 2, moderate; and 3, strong staining. Cases showing moderate (score 2) or strong (score 3) staining in more than 10% were regarded as “high expression.” Cases with no or weak staining were regarded as “low expression.” This assessment was performed by two pathologists blind to all clinicopathologic data and outcome variables. Due to tissue loss in microarray cores, three samples of SIRT7 (n=275) and two of H3K18ac (n=276) were excluded. None of the ELK4 (n=278) samples were excluded.
- Statistical analysis
- To examine univariate overall survival, the log-rank test with the Kaplan-Meier survival curve was used. The Cox’s proportional hazards regression model was used for univariate and multivariate analyses of overall and disease-free survival. Overall survival rate was calculated from the date of surgery to the date of the latest follow-up or death. The disease-free survival rate was also calculated from the date of surgery to the date of recurrence on follow-up images or pathologic confirmation. For comparing means, the independent t test was used. Pearson’s correlation coefficient was applied to the correlation between IHC expression and the clinicopathologic data. The p-value less than .05 was considered to be statistically significant. All statistical analyses were performed using the SPSS ver. 20 (IBM Corp., Armonk, NY, USA).
MATERIALS AND METHODS
- Patient characteristics
- In the present study, there were 231 (83%) male and 47 (17%) female HCC patients between 26–84 years of age (median±standard deviation, 57±10.2 years) who underwent curative operation. The median postoperational follow-up period was 34 months. A total of 56 patients (20%) died during the follow-up period. Among all patients, 149 (54%) were in AJCC stage I, 95 (34%) were in stage II, nine (3%) were in stage III, and 25 (9%) were in stage IV.
- Expressions of SIRT7, H3K18ac, and ELK4 in non-tumor and tumor cells
- The results of all three antibodies showed significant nuclear expression patterns (Fig. 1). In non-tumor hepatocytes of 52 patients, high SIRT7, H3K18ac, and ELK4 expressions were observed in seven (13%), four (8%), and one (2%) patients, respectively; however, in tumor cells, 51 (19%), 143 (52%), and 66 (24%) patients showed high expressions for SIRT7, H3K18ac, and ELK4, respectively (Table 1). Interestingly, H3K18ac showed significantly high expression in tumor cells (mean value, 1.08 and 1.71, respectively; p<.001). In addition, SIRT7 was higher in tumor cells (mean value, 1.06 and 1.08, respectively; p=.858) and ELK4 was higher in non-tumor hepatocytes (mean value, 0.96 and 0.87, respectively; p=.120). However, these results were not significant.
- Analyses of overall and disease-free survival
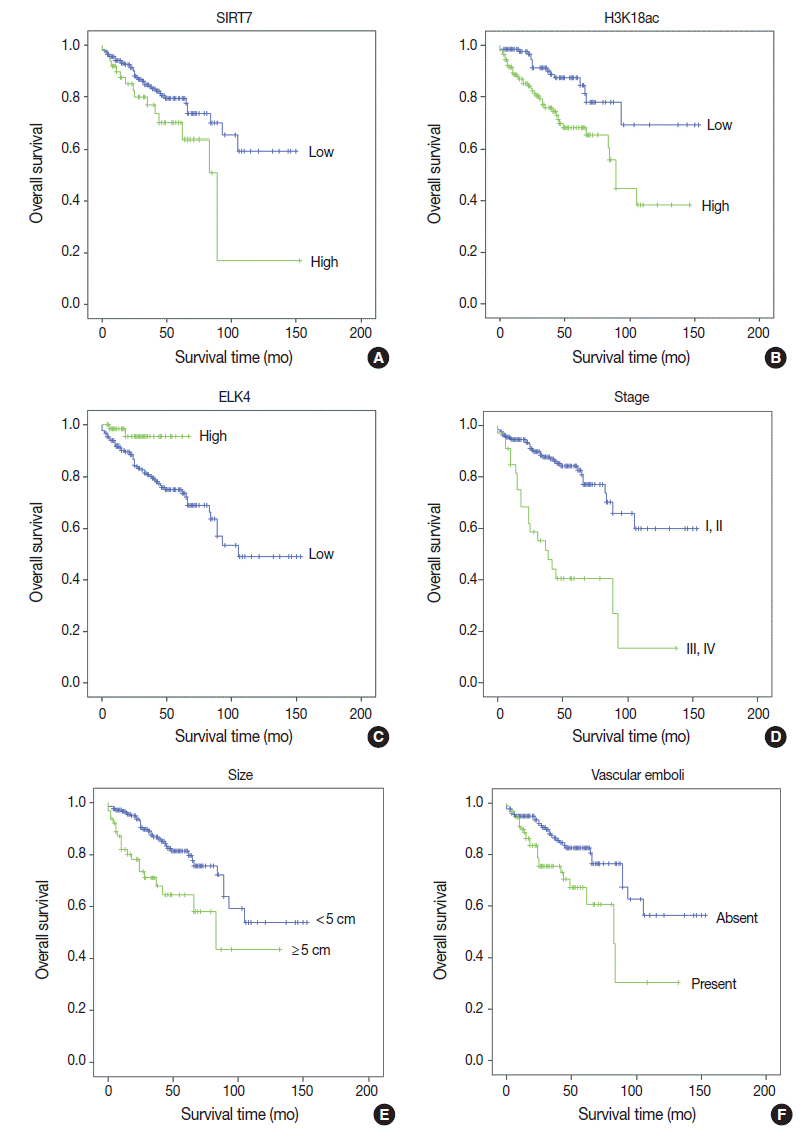
- Univariate analyses of overall survival rate using the Kaplan-Meier method showed that high expression in both SIRT7 and H3K18ac were associated with poor overall survival rate (p=.034 and p=.001, respectively) (Fig. 2A, B); however, high expression in ELK4 was associated with good overall survival (p=.021) (Fig. 2C). High AJCC stage (p<.001) (Fig. 2D), large tumor size (≥5 cm) (p=.001) (Fig. 2E), and the presence of vascular emboli (p=.003) (Fig. 2F) were associated with poor overall survival rate. Additionally, younger age (<65 years) (p=.028) and positive surgical margin (p=.001) were correlated with poor outcomes. Analyses using Cox’s proportional hazards are shown in Table 2.
- In multivariate analyses (Table 2), high expression in H3K18ac was determined to be a poor prognostic factor (p=.001), whereas high expression in ELK4 was indicated as a good prognostic factor (p=.015). However, high SIRT7 expression was unremarkable (p=.368). Tumor size ≥5 cm, positive margin, and advanced stage (III–IV) (p=.024, p=.012, and p<.001, respectively) were related independently as poor prognostic factors. Finally, the presence of vascular emboli was not significant (p=.409).
- Disease-free survival rate analyses were performed using the Cox’s proportional hazard model. High SIRT7 expression, presence of vascular emboli, AJCC stage III-IV and Edmondson-Steiner grade III–IV were associated with worse prognosis (Table 3). However, only AJCC stage was shown to be an independent factor in the multivariate analysis (p=.021).
- Analyses of correlation
- SIRT7 and H3K18ac showed a positive correlation (p=.006) (Table 4); however, ELK4 did not show a significant correlation with the other two markers. Tumor size showed a positive association with AJCC stage (p<.001), vascular tumor emboli (p=.001), and necrosis (p<.001). Additionally, tumor necrosis was positively associated with presence of vascular emboli (p=.012) as well as AJCC stage III and IV (p=.005). Edmondson-Steiner grade III and IV showed positive correlation with the existence of vascular emboli (p=.000) and high AJCC stage (p=.010). Elder patients (≥65 years) had positive association with positive surgical margin (p=.029).
- Table 1 summarizes the correlation between IHC markers and clinicopathologic data. Edmondson-Steiner grade III and IV had a correlation with high SIRT7 and high H3K18ac expression (p=.002 and p<.001, respectively). A high T stage or presence of vascular emboli were associated with high H3K18ac expression (p=.027 and p=.012, respectively). In addition, high H3K18ac expression showed a significant correlation with females (p=.033) and high ELK4 expression with males (p=.021).
RESULTS
- The objective of the present study was to analyze H3K18ac, which is deacetylated by SIRT7 linked with ELK4, by IHC expression in HCC samples. We hypothesized that high IHC expression of SIRT7 would be associated with high ELK4 and low H3K18ac expression, and these results would lead to a poor overall survival rate. On the contrary to the hypothesis, this study found that SIRT7 expression had a weak positive correlation with H3K18ac, but no correlation with ELK4 (Table 4). In addition, high H3K18ac expression was unexpectedly associated with poor prognosis, whereas high ELK4 expression was associated with better prognosis in the overall survival analyses (Table 2). Lastly, high SIRT7 expression associated with poor prognosis was in accordance with our major hypothesis.
- There is no definite explanation of the unexpected positive correlation of SIRT7 and H3K18ac, but there are several possible mechanisms. First, other pathways may exist. For example, SIRT2 and SIRT6 can also deacetylate H3K18ac in addition to SIRT7 [18,19]. Conversely, SIRT7 also deacetyl-ates p53 in addition to H3K18ac [20]. Although the prognostic direction of H3K18ac was unexpected, it seems to have significance, which is shown in the univariate and multivariate overall survival analyses. In addition, correlation analyses revealed a positive association with poor Edmondson-Steiner grade, high T stage and presence of vascular emboli (Tables 1, 2). Putting these evidences together, we can suggest that there are other important mechanisms beyond the deacetylation of H3K18ac by SIRT7. The weak correlation coefficient value may be a reflection of this (Table 4).
- Second, the method of approach was different. In previous studies, experiments were performed with cell-lines and were focused on the specific gene foci [12,14]. In the present study, we used surgical specimens and examined overall protein expressions by the IHC approach. Some authors have argued that gene-specific expression of histone acetylation can run contrary to the overall state of modification. This phenomenon could have resulted from compensation and made a difference in the overall expression of acetylation seen in the IHC study [21-23]. Further evaluation of the relationship of SIRT7 and H3K18ac in HCC is needed [19]. Because previous HCC studies of H3K18ac were focused on tumor suppressor genes, the roles of H3K18ac in proto-oncogene or global H3K18ac expression in HCC progression have not been sufficiently investigated. However, several studies have shown the correlation of H3K18ac with poor prognosis in other malignancies, such as pancreatic adenocarcinoma or esophageal cancer [23,24]. Moreover, in some malignant tumors, especially in HCCs, global acetylation of H3K18 could be more associated with genes related to tumor progression or proliferation than tumor suppression [25-27].
- High SIRT7 expression in this study was associated with poor overall survival rate and high tumor grade but did not show a statistical correlation with H3K18ac and ELK4 expression. Most studies on SIRT7 in HCC have focused on the HDAC activity of SIRT7 in relation to tumor suppressor genes [12,14]. Recent studies have revealed other mechanisms of SIRT7 in human cells. They have reported that SIRT7 can regulate protein folding in mitochondria, and a decrease in SIRT7 could result in stem cell senescence [28]. Another study also reported that SIRT7 can interact with SIRT1, resulting in epithelial-to-mesenchymal transition via control of E-cadherin. The mechanism of SIRT7 as a poor prognostic factor in HCC may be more related to other roles in tumor cells than its role as a suppressor of tumor suppressor genes [29].
- High ELK4 expression did not show a correlation with SIRT7 or H3K18ac in the present study. However, high ELK4 expression was an independent factor of good prognosis in HCC patients. The most studied mechanism of ELK4 in tumorigenesis is the role as a cofactor of SIRT7 in deacetylation of H3K18, which is located in tumor suppressor genes or ribosome-related genes. Additionally, ELK4 fundamentally forms complexes with SRF dimers on SREs found in the c-fos and other immediate early gene promoters [15], and this process is stimulated by the activated mitogen-activated protein kinase (MAPK) signaling pathway [15,30]. Therefore, we can assume that high ELK4 status would induce the oncogenic pathway, but this has not been investigated. Furthermore, ELK4 is expected to inhibit gene transcription, due to its resemblance to ELK3/NET. ELK3/NET acts as a transcription repressor of target promoters through its NET inhibitory domain (NID) and C-terminal-binding protein inhibition domain in the absence of the MAPK signal [31,32]. Thus, loss of ELK3 expression has been reported in malignant mesothelioma, suggesting the role of ELK3 as a repressor of tumors [33]. ELK4 also has a similar NID region to ELK3/NET [34,35], suggesting that ELK4 may also inhibit gene transcription during the inactivated MAPK condition [36]. The role of ELK4 in HCC requires more investigation, but this transcriptional repression could be associated with inhibition of tumor progression.
- In conclusion, high expression of ELK4 and low expression of H3K18ac were independent and good prognostic factors in HCC patients. High SIRT7 expression in HCC was associated with poor overall survival rate, although it was not an independent prognostic factor. In HCC tissue samples, these proteins had significant prognostic values independently; however, we did not find an integrated tumorigenic effect. Since these proteins have not been widely investigated, more comprehensive studies for interactions and mechanisms of ELK4, SIRT7, and H3K18ac are needed in HCC.
DISCUSSION
-
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Notes
Fig. 1.Characteristic nuclear staining of tumor cells by immunohistochemistry. Low SIRT7 expression (A), high SIRT7 expression (B), low H3K18ac expression (C), high H3K18ac expression (D), low ELK4 expression (E), and high ELK4 expression (F) are seen.
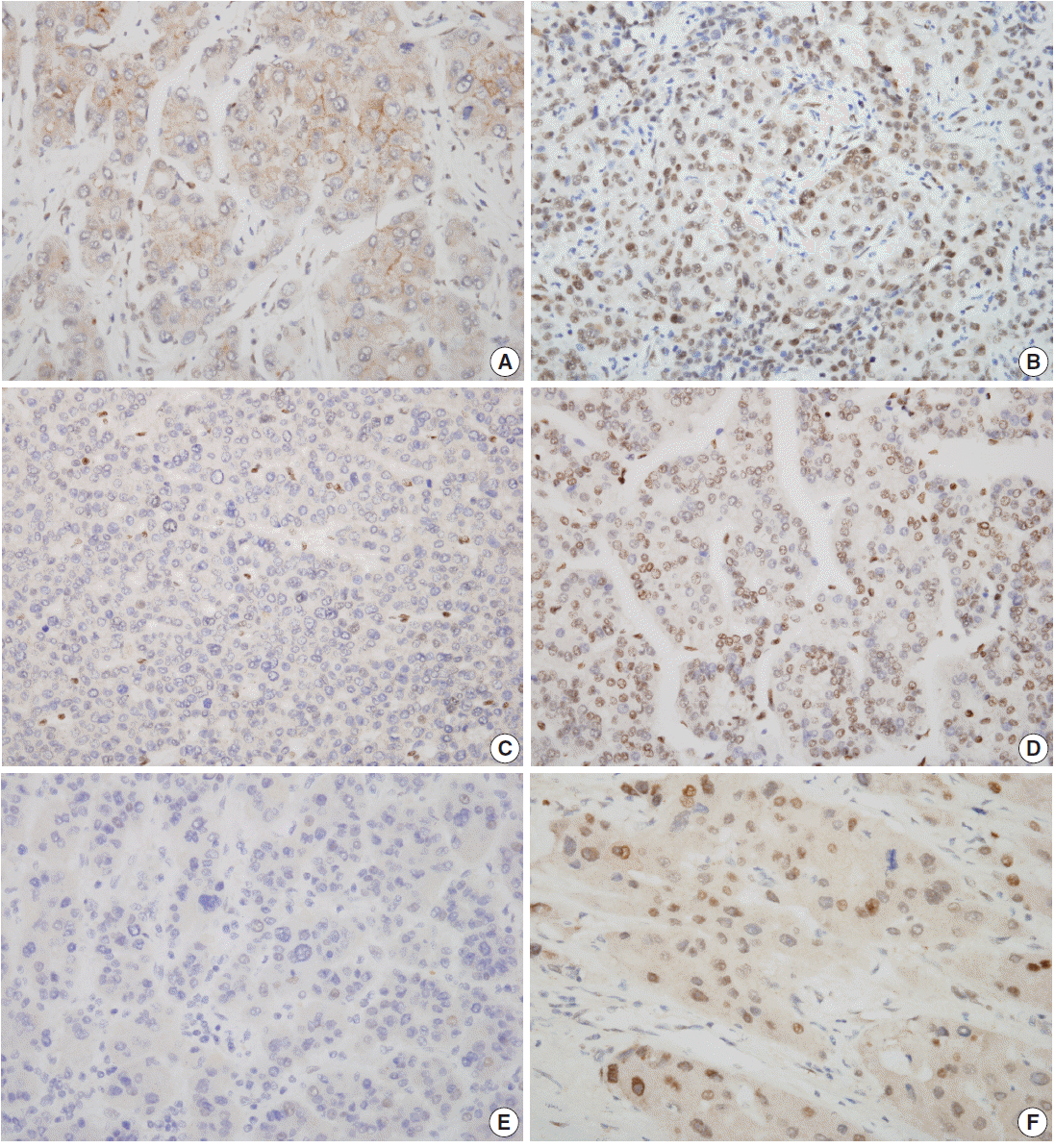

Fig. 2.Kaplan-Meier survival curves of immunohistochemical (IHC) markers and clinicopathologic data. With IHC markers (A–C), high SIRT7 expression (A) and high H3K18ac expression (B) were associated with poor prognosis (p = .034 and p = .001, respectively). However, high ETS-like transcription factor 4 (ELK4) expression was associated with good prognosis (p = .021) (C). With clinicopathologic factors (D–F), high American Joint Committee on Cancer stage (III, IV) (p < .001) (D), large tumor size (maximal diameter ≥ 5 cm) (p = .001) (E), and the presence of vascular emboli (F) were associated with poor overall survival (p = .003).
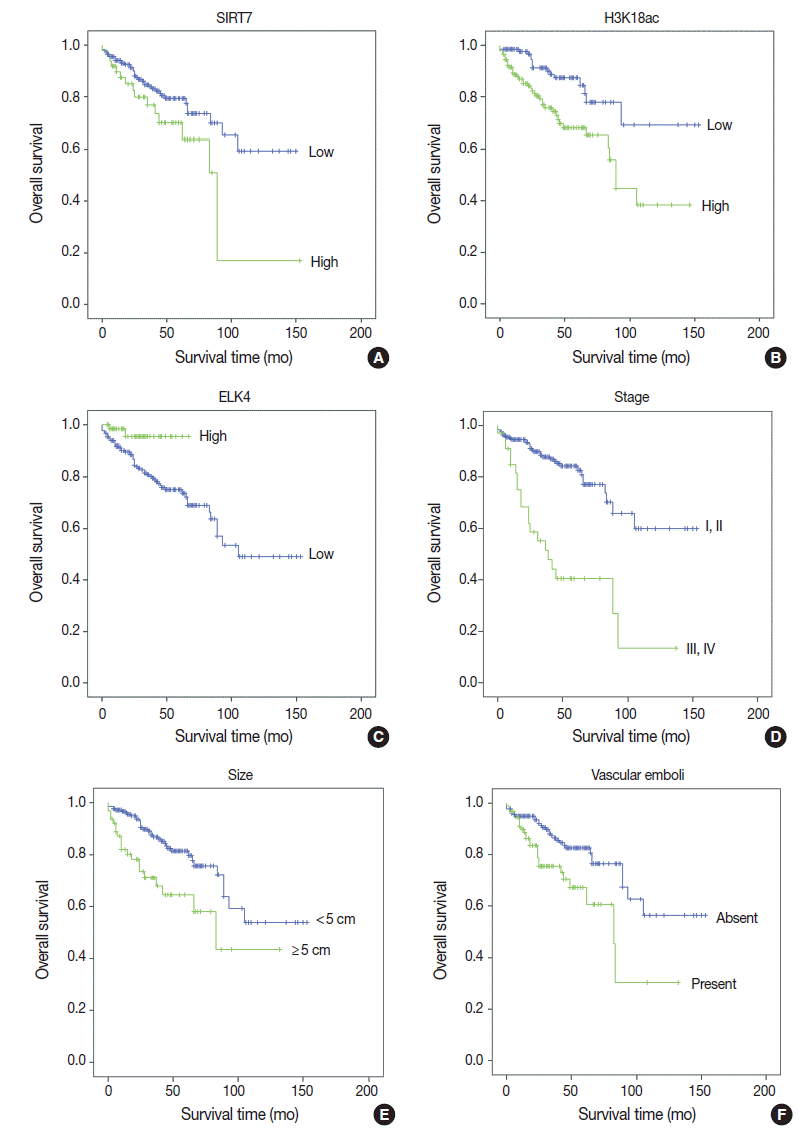

Table 1.Correlation between immunohistochemical markers and clinicopathologic data
| Characteristic | No. (%) |
Immunohistochemistry expression |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
SIRT7 (n = 275) |
H3K18Ac (n = 276) |
ELK4 (n = 278) |
||||||||
| Low (n = 224, 81%) | High (n = 51, 19%) | p-value | Low (n = 133, 48%) | High (n = 143, 52%) | p-value | Low (n = 222, 76%) | High (n = 66, 24%) | p-value | ||
| Age (yr) | ||||||||||
| < 65 | 214 (77) | 176 | 37 | .721 | 80 | 115 | .184 | 80 | 115 | .184 |
| ≥ 65 | 64 (23) | 50 | 12 | 35 | 28 | 35 | 28 | |||
| Sex | ||||||||||
| Female | 47 (16.9) | 36 | 9 | .448 | 16 | 31 | .021* | 16 | 31 | .033* |
| Male | 231 (83) | 190 | 40 | 117 | 112 | 117 | 112 | |||
| Edmondson-Steiner grade | ||||||||||
| I–II | 178 (64) | 154 | 23 | .002* | 107 | 76 | < .001* | 135 | 43 | .828 |
| III–IV | 100 (36) | 72 | 26 | 26 | 67 | 77 | 23 | |||
| Tumor size (cm) | ||||||||||
| < 5 | 214 (77) | 176 | 37 | .721 | 103 | 111 | .972 | 163 | 51 | .948 |
| ≥ 5 | 64 (23) | 50 | 12 | 30 | 32 | 49 | 15 | |||
| AJCC stage | ||||||||||
| I–II | 244 (87.8) | 199 | 42 | .653 | 119 | 123 | .384 | 183 | 61 | .188 |
| III–IV | 34 (12.2) | 27 | 7 | 14 | 20 | 29 | 5 | |||
| Vascular emboli | ||||||||||
| Absent | 183 (65.8) | 153 | 29 | .255 | 98 | 85 | .012* | 148 | 35 | .056 |
| Present | 95 (34.2) | 73 | 20 | 35 | 58 | 64 | 29 | |||
| Margin | ||||||||||
| Negative | 263 (94.6) | 214 | 46 | .821 | 125 | 136 | .683 | 199 | 64 | .727 |
| Positive | 15 (5.4) | 12 | 3 | 8 | 7 | 13 | 2 | |||
| Tumor necrosis | ||||||||||
| Absent | 158 (61.7) | 131 | 25 | .261 | 80 | 77 | .133 | 113 | 45 | .211 |
| Present | 98 (38.3) | 76 | 21 | 40 | 57 | 77 | 21 | |||
Table 2.Univariate and multivariate analyses of overall survival using Cox’s proportional hazards model
| Variable |
Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| High SIRT7 expression | 1.853 (1.036–3.313) | .038* | 1.323 (0.720–2.430) | .368 |
| High H3K18Ac expression | 2.569 (1.438–4.592) | .001* | 2.731 (1.469–5.076) | .001* |
| High ELK4 expression | 0.219 (0.053–0.907) | .036* | 0.162 (0.037–0.704) | .015* |
| Tumor size (≥ 5 cm) | 2.427 (1.400–4.207) | .002* | 2.044 (1.104–3.787) | .023* |
| Vascular emboli | 2.176 (1.273–3.720) | .004* | 1.287 (0.707–2.343) | .409 |
| Margin positive | 3.371 (1.582–7.183) | .002* | 2.943 (1.300–6.661) | .010* |
| AJCC stage III–IV | 4.302 (2.489–7.438) | < .001* | 3.007 (1.650–5.483) | < .001* |
Table 3.Univariate and multivariate analysis using Cox’s proportional hazards analysis of disease-free survival
| Variable |
Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| High SIRT7 expression | 1.573 (1.013–2.444) | .044* | 1.440 (0.912–2.276) | .118 |
| High H3K18Ac expression | 1.229 (0.844–1.789) | .283 | 1.022 (0.680–1.537) | .915 |
| High ELK4 expression | 0.922 (0.566–1.502) | .744 | 0.901 (0.545–1.490) | .685 |
| Tumor size (≥ 5 cm) | 1.393 (0.903–2.150) | .134 | 1.110 (0.697–1.769) | .660 |
| Vascular emboli | 1.465 (1.001–2.144) | .049* | 1.192 (0.790–1.800) | .402 |
| Margin positive | 1.200 (0.558–2.580) | .640 | 0.903 (0.403–2.025) | .805 |
| AJCC stage III–IV | 2.128 (1.310–3.458) | .002* | 1.898 (1.101–3.273) | .021* |
| Edmondson-steiner’s grade | 1.513 (1.040–2.202) | .030* | 1.270 (0.839–1.923) | .258 |
Table 4.Bivariate analysis of SIRT7, H3K18ac, and ELK4 (n = 275)
| SIRT7 | H3K18ac | ELK4 | ||
|---|---|---|---|---|
| SIRT7 | Correlation coefficient | 1.000 | 0.165 | –0.054 |
| p-value | .006* | .372 | ||
| H3K18ac | Correlation coefficient | 0.165 | 1.000 | –0.035 |
| p-value | .006* | .560 | ||
| ELK4 | Correlation coefficient | –0.054 | –0.035 | 1.000 |
| p-value | .372 | .560 |
- 1. Jung KW, Won YJ, Kong HJ, Oh CM, Lee DH, Lee JS. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2011. Cancer Res Treat 2014; 46: 109–23. ArticlePubMedPMCPDF
- 2. Kim HS, Shen Q, Nam SW. Histone deacetylases and their regulatory microRNAs in hepatocarcinogenesis. J Korean Med Sci 2015; 30: 1375–80. ArticlePubMedPMC
- 3. Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol 2014; 20: 333–45. ArticlePubMedPMC
- 4. Hung SY, Lin HH, Yeh KT, Chang JG. Histone-modifying genes as biomarkers in hepatocellular carcinoma. Int J Clin Exp Pathol 2014; 7: 2496–507. PubMedPMC
- 5. Li L, Bhatia R. The controversial role of sirtuins in tumorigenesis: SIRT7 joins the debate. Cell Res 2013; 23: 10–2. ArticlePubMed
- 6. Mellini P, Valente S, Mai A. Sirtuin modulators: an updated patent review (2012 - 2014). Expert Opin Ther Pat 2015; 25: 5–15. ArticlePubMed
- 7. Kleszcz R, Paluszczak J, Baer-Dubowska W. Targeting aberrant cancer metabolism: the role of sirtuins. Pharmacol Rep 2015; 67: 1068–80. ArticlePubMed
- 8. Jang KY, Noh SJ, Lehwald N, et al. SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS One 2012; 7: e45119. Article
- 9. Chen J, Chan AW, To KF, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology 2013; 57: 2287–98. ArticlePubMed
- 10. Zhang B, Qin L, Zhou CJ, Liu YL, Qian HX, He SB. SIRT3 expression in hepatocellular carcinoma and its impact on proliferation and invasion of hepatoma cells. Asian Pac J Trop Med 2013; 6: 649–52. ArticlePubMed
- 11. Zhang ZG, Qin CY. Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signalregulated kinase signaling pathway. Mol Med Rep 2014; 9: 882–8. ArticlePubMed
- 12. Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology 2013; 57: 1055–67. ArticlePubMed
- 13. Zheng Y, Chen H, Yin M, et al. MiR-376a and histone deacetylation 9 form a regulatory circuitry in hepatocellular carcinoma. Cell Physiol Biochem 2015; 35: 729–39. ArticlePubMedPDF
- 14. Barber MF, Michishita-Kioi E, Xi Y, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012; 487: 114–8. ArticlePubMedPMC
- 15. Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene 2004; 324: 1–14. ArticlePubMed
- 16. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC cancer staging manual. New York: Springer, 2010; 7th.
- 17. Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 1954; 7: 462–503. ArticlePubMed
- 18. Eskandarian HA, Impens F, Nahori MA, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 2013; 341: 1238858.ArticlePubMed
- 19. Tasselli L, Xi Y, Zheng W, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol 2016; 23: 434–40. ArticlePubMedPMC
- 20. Vakhrusheva O, Smolka C, Gajawada P, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 2008; 102: 703–10. ArticlePubMed
- 21. Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005; 122: 517–27. ArticlePubMed
- 22. Sinha I, Wirén M, Ekwall K. Genome-wide patterns of histone modifications in fission yeast. Chromosome Res 2006; 14: 95–105. ArticlePubMed
- 23. Juliano CN, Izetti P, Pereira MP, et al. H4K12 and H3K18 acetylation associates with poor prognosis in pancreatic cancer. Appl Immunohistochem Mol Morphol 2016; 24: 337–44. ArticlePubMed
- 24. Tzao C, Tung HJ, Jin JS, et al. Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod Pathol 2009; 22: 252–60. ArticlePubMed
- 25. Liu BL, Cheng JX, Zhang X, et al. Global histone modification patterns as prognostic markers to classify glioma patients. Cancer Epidemiol Biomarkers Prev 2010; 19: 2888–96. ArticlePubMed
- 26. Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005; 435: 1262–6. ArticlePubMed
- 27. Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 2009; 174: 1619–28. ArticlePubMedPMC
- 28. Liu JP, Chen R. Stressed SIRT7: facing a crossroad of senescence and immortality. Clin Exp Pharmacol Physiol 2015; 42: 567–9. ArticlePubMed
- 29. Malik S, Villanova L, Tanaka S, et al. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci Rep 2015; 5: 9841.ArticlePubMedPMC
- 30. Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2001; 2: 827–37. ArticlePubMed
- 31. Criqui-Filipe P, Ducret C, Maira SM, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and deacetylation. EMBO J 1999; 18: 3392–403. ArticlePubMedPMC
- 32. Maira SM, Wurtz JM, Wasylyk B. Net (ERP/SAP2) one of the Rasinducible TCFs, has a novel inhibitory domain with resemblance to the helix-loop-helix motif. EMBO J 1996; 15: 5849–65. ArticlePubMedPMCPDF
- 33. van Riggelen J, Buchwalter G, Soto U, et al. Loss of net as repressor leads to constitutive increased c-fos transcription in cervical cancer cells. J Biol Chem 2005; 280: 3286–94. ArticlePubMed
- 34. Stinson J, Inoue T, Yates P, Clancy A, Norton JD, Sharrocks AD. Regulation of TCF ETS-domain transcription factors by helix-loophelix motifs. Nucleic Acids Res 2003; 31: 4717–28. ArticlePubMedPMC
- 35. Yates PR, Atherton GT, Deed RW, Norton JD, Sharrocks AD. Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J 1999; 18: 968–76. ArticlePubMedPMC
- 36. Kaikkonen S, Makkonen H, Rytinki M, Palvimo JJ. SUMOylation can regulate the activity of ETS-like transcription factor 4. Biochim Biophys Acta 2010; 1799: 555–60. ArticlePubMed
References
Figure & Data
References
Citations
Citations to this article as recorded by 

- SIRT7: the seventh key to unlocking the mystery of aging
Umar Raza, Xiaolong Tang, Zuojun Liu, Baohua Liu
Physiological Reviews.2024; 104(1): 253. CrossRef - The Significance of Modified Histone H3 in Epithelial Dysplasia and Oral Cancer
Woraphaluck Tachaveeraphong, Ekarat Phattarataratip
International Dental Journal.2024;[Epub] CrossRef - Role of Sirtuins in the Pathogenesis of Rheumatoid Arthritis
Agata Poniewierska-Baran, Oliwia Bochniak, Paulina Warias, Andrzej Pawlik
International Journal of Molecular Sciences.2023; 24(2): 1532. CrossRef - Role of sirtuins in hepatocellular carcinoma progression and multidrug resistance: Mechanistical and pharmacological perspectives
María Paula Ceballos, Ariel Darío Quiroga, Nicolás Francisco Palma
Biochemical Pharmacology.2023; 212: 115573. CrossRef - Substrates and Cyclic Peptide Inhibitors of the Oligonucleotide‐Activated Sirtuin 7**
Julie E. Bolding, Alexander L. Nielsen, Iben Jensen, Tobias N. Hansen, Line A. Ryberg, Samuel T. Jameson, Pernille Harris, Günther H. J. Peters, John M. Denu, Joseph M. Rogers, Christian A. Olsen
Angewandte Chemie International Edition.2023;[Epub] CrossRef - Substrates and Cyclic Peptide Inhibitors of the Oligonucleotide‐Activated Sirtuin 7**
Julie E. Bolding, Alexander L. Nielsen, Iben Jensen, Tobias N. Hansen, Line A. Ryberg, Samuel T. Jameson, Pernille Harris, Günther H. J. Peters, John M. Denu, Joseph M. Rogers, Christian A. Olsen
Angewandte Chemie.2023;[Epub] CrossRef - Epigenomic interplay in tumor heterogeneity: Potential of epidrugs as adjunct therapy
Suvasmita Rath, Diptesh Chakraborty, Jyotsnarani Pradhan, Mohammad Imran Khan, Jagneshwar Dandapat
Cytokine.2022; 157: 155967. CrossRef - Distinct histone H3 modification profiles correlate with aggressive characteristics of salivary gland neoplasms
Aroonwan Lam-Ubol, Ekarat Phattarataratip
Scientific Reports.2022;[Epub] CrossRef - Acetyl-CoA: An interplay between metabolism and epigenetics in cancer
Yang Hao, Qin Yi, Xu XiaoWu, Chen WeiBo, Zu GuangChen, Chen XueMin
Frontiers in Molecular Medicine.2022;[Epub] CrossRef - Sirtuins (SIRTs) As a Novel Target in Gastric Cancer
Agata Poniewierska-Baran, Paulina Warias, Katarzyna Zgutka
International Journal of Molecular Sciences.2022; 23(23): 15119. CrossRef - Novel oncogenes and tumor suppressor genes in hepatocellular carcinoma
Fang Wang, Peter Breslin S J, Wei Qiu
Liver Research.2021; 5(4): 195. CrossRef - Acute high folic acid treatment in SH-SY5Y cells with and without MTHFR function leads to gene expression changes in epigenetic modifying enzymes, changes in epigenetic marks, and changes in dendritic spine densities
Daniel F. Clark, Rachael Schmelz, Nicole Rogers, Nuri E. Smith, Kimberly R. Shorter, Lorenzo Chiariotti
PLOS ONE.2021; 16(1): e0245005. CrossRef - The E-Twenty-Six Family in Hepatocellular Carcinoma: Moving into the Spotlight
Tongyue Zhang, Danfei Liu, Yijun Wang, Mengyu Sun, Limin Xia
Frontiers in Oncology.2021;[Epub] CrossRef - Upregulation of histone acetylation reverses organic anion transporter 2 repression and enhances 5-fluorouracil sensitivity in hepatocellular carcinoma
Yingying Wang, Qianying Zhu, Haihong Hu, Hong Zhu, Bo Yang, Qiaojun He, Lushan Yu, Su Zeng
Biochemical Pharmacology.2021; 188: 114546. CrossRef - HCG11 up-regulation induced by ELK4 suppressed proliferation in vestibular schwannoma by targeting miR-620/ELK4
Ruiqing Long, Zhuohui Liu, Jinghui Li, Yuan Zhang, Hualin Yu
Cancer Cell International.2021;[Epub] CrossRef - Downregulation of circular RNA circPVT1 restricts cell growth of hepatocellular carcinoma through downregulation of Sirtuin 7 via microRNA‐3666
Yong Li, Haitao Shi, Jia Yuan, Lu Qiao, Lei Dong, Yan Wang
Clinical and Experimental Pharmacology and Physiology.2020; 47(7): 1291. CrossRef - Clinicopathological and molecular analysis of SIRT7 in hepatocellular carcinoma
Masae Yanai, Morito Kurata, Yutaka Muto, Hiroto Iha, Toshinori Kanao, Anna Tatsuzawa, Sachiko Ishibashi, Masumi Ikeda, Masanobu Kitagawa, Kouhei Yamamoto
Pathology.2020; 52(5): 529. CrossRef - MicroRNA-148b Inhibits the Malignant Biological Behavior of Melanoma by Reducing Sirtuin 7 Expression Levels
Rui Sun, Meiliang Guo, Xiaojing Fan, Qinqin Meng, Dingfen Yuan, Xinrong Yang, Kexiang Yan, Hui Deng, Fengjie Sun
BioMed Research International.2020; 2020: 1. CrossRef - H3K18Ac as a Marker of Cancer Progression and Potential Target of Anti-Cancer Therapy
Marta Hałasa, Anna Wawruszak, Alicja Przybyszewska, Anna Jaruga, Małgorzata Guz, Joanna Kałafut, Andrzej Stepulak, Marek Cybulski
Cells.2019; 8(5): 485. CrossRef - Sirtuin7 has an oncogenic potential via promoting the growth of cholangiocarcinoma cells
Wenzhi Li, Zhe Sun, Chen Chen, Lin Wang, Zhimin Geng, Jie Tao
Biomedicine & Pharmacotherapy.2018; 100: 257. CrossRef - Identification of cancer‐related potential biomarkers based on lncRNA–pseudogene–mRNA competitive networks
Cheng Wu, Yunzhen Wei, Yinling Zhu, Kun Li, Yanjiao Zhu, Yichuan Zhao, Zhiqiang Chang, Yan Xu
FEBS Letters.2018; 592(6): 973. CrossRef - SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation
Wenlu Li, Dandan Zhu, Shuaihua Qin
Journal of Experimental & Clinical Cancer Research.2018;[Epub] CrossRef - Sirtuin 7: a new marker of aggressiveness in prostate cancer
Romain Haider, Fabienne Massa, Lisa Kaminski, Stephan Clavel, Zied Djabari, Guillaume Robert, Kathiane Laurent, Jean-François Michiels, Matthieu Durand, Jean-Ehrland Ricci, Jean-François Tanti, Frédéric Bost, Damien Ambrosetti
Oncotarget.2017; 8(44): 77309. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure

 E-submission
E-submission